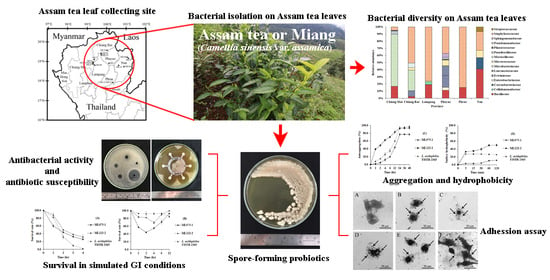
Culturable Bacterial Community on Leaves of Assam Tea (Camellia sinensis var. assamica) in Thailand and Human Probiotic Potential of Isolated Bacillus spp.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Site and Collection of Assam Tea Leaves
2.2. Isolation of Bacteria
2.3. Bacterial Genomic DNA Extraction
2.4. 16S rRNA Gene Amplification
2.5. Probiotic Property Characterization of Bacillus Strains
2.5.1. Antibacterial Activity Investigation
2.5.2. Antibiotic Susceptibility Assay
2.5.3. Survival Evaluation in Simulated Gastric and Intestinal Fluids
2.5.4. Cellular Autoaggregation Assay
2.5.5. Cell Surface Hydrophobicity Assay
2.5.6. In Vitro Bacterial Adhesion Assay
2.6. Statistical Analysis
3. Results
3.1. Bacterial Isolation from Assam Tea Leaves and 16S rRNA Gene Identification
3.2. Bacterial Community on Assam Tea Leaf Surface
3.3. Bacterial Alpha Diversity, Richness, and Community Structure
3.4. Probiotic Property Determination of Bacillus Strains
3.4.1. Antibacterial Activity
3.4.2. Antibiotic Susceptibility
3.4.3. Survival in Gastrointestinal Tract Conditions, Autoaggregation, and Cell Surface Hydrophobicity Investigation
3.4.4. Bacterial Adhesion
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Okada, S.; Daengsubha, W.; Uchimura, T.A.I.; Ohara, N.; Kozaki, M. Flora of lactic acid bacteria in miang produced in northern Thailand. J. Gen. Appl. Microbiol. 1986, 32, 57–65. [Google Scholar] [CrossRef]
- Kanpiengjai, A.; Chui-Chai, N.; Chaikaew, S.; Khanongnuch, C. Distribution of tannin-’tolerant yeasts isolated from Miang, a traditional fermented tea leaf (Camellia sinensis var. assamica) in northern Thailand. Int. J. Food Microbiol. 2016, 238, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Khanongnuch, C.; Unban, K.; Kanpiengjai, A.; Saenjum, C. Recent research advances and ethno-botanical history of miang, a traditional fermented tea (Camellia sinensis var. assamica) of northern Thailand. J. Ethn. Foods 2017, 4, 135–144. [Google Scholar] [CrossRef]
- Tanasupawat, S.; Pakdeeto, A.; Thawai, C.; Yukphan, P.; Okada, S. Identification of lactic acid bacteria from fermented tea leaves (miang) in Thailand and proposals of Lactobacillus thailandensis sp. nov., Lactobacillus camelliae sp. nov., and Pediococcus siamensis sp. nov. J. Gen. Appl. Microbiol. 2007, 53, 7–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaikaew, S.; Baipong, S.; Sone, T.; Kanpiengjai, A.; Chui-chai, N.; Asano, K.; Khanongnuch, C. Diversity of lactic acid bacteria from Miang, a traditional fermented tea leaf in northern Thailand and their tannin-tolerant ability in tea extract. J. Microbiol. 2017, 55, 720–729. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Kemmitt, S.J.; Brookes, P.C. Soil microbial biomass and activity in Chinese tea gardens of varying stand age and productivity. Soil Biol. Biochem. 2007, 39, 1468–1478. [Google Scholar] [CrossRef]
- Li, Y.C.; Li, Z.; Li, Z.W.; Jiang, Y.H.; Weng, B.Q.; Lin, W.X. Variations of rhizosphere bacterial communities in tea (Camellia sinensis L.) continuous cropping soil by high-throughput pyrosequencing approach. J. Appl. Microbiol. 2016, 121, 787–799. [Google Scholar] [CrossRef]
- Zhang, Y.; Skaar, I.; Sulyok, M.; Liu, X.; Rao, M.; Taylor, J.W. The microbiome and metabolites in fermented Pu-erh tea as revealed by high-throughput sequencing and quantitative multiplex metabolite analysis. PLoS ONE 2016, 11, e0157847. [Google Scholar] [CrossRef]
- Nancib, A.; Nancib, N.; Boudrant, J. Production of lactic acid from date juice extract with free cells of single and mixed cultures of Lactobacillus casei and Lactococcus lactis. World J. Microbiol. Biotechnol. 2009, 25, 1423–1429. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Ripple, W.J.; Timmis, K.N.; Azam, F.; Bakken, L.R.; Baylis, M.; Behrenfeld, M.J.; Boetius, A.; Boyd, P.W.; Classen, A.T.; et al. Scientists’ warning to humanity: Microorganisms and climate change. Nat. Rev. Microbiol. 2019, 17, 569–586. [Google Scholar] [CrossRef] [Green Version]
- Rungsirivanich, P.; Inta, A.; Tragoolpua, Y.; Thongwai, N. Partial rpoB gene sequencing identification and probiotic potential of Floricoccus penangensis ML061-4 isolated from Assam tea (Camellia sinensis var. assamica). Sci. Rep. 2019, 9, 16561. [Google Scholar] [CrossRef] [PubMed]
- Rungsirivanich, P.; Thongwai, N. Antibacterial activity and tannin tolerance of Bacillus spp. isolated from leaves of Miang (Camellia sinensis (L.) Kuntze var. assamica (J.W. Mast.) Kitam.). Int. J. Biosci. Biochem. Bioinform. 2020, 10, 26–33. [Google Scholar] [CrossRef] [Green Version]
- FAO/WHO. Report of a Joint Food and Agriculture Organization of the United Nations and World Health Organization (FAO/WHO) expert consultation on guidelines for the evaluation of probiotics in food. Available online: http://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf (accessed on 5 August 2002).
- Angmo, K.; Kumari, A.; Savitri; Bhalla, T.C. Probiotic characterization of lactic acid bacteria isolated from fermented foods and beverage of Ladakh. LWT Food Sci. Technol. 2016, 66, 428–435. [Google Scholar] [CrossRef]
- Meidong, R.; Doolgindachbaporn, S.; Jamjan, W.; Sakai, K.; Tashiro, Y.; Okugawa, Y.; Tongpim, S. A novel probiotic Bacillus siamensis B44v isolated from Thai pickled vegetables (Phak-dong) for potential use as a feed supplement in aquaculture. J. Gen. Appl. Microbiol. 2017, 63, 246–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abriouel, H.; Franz, C.M.A.P.; Omar, N.B.; Gálvez, A. Diversity and applications of Bacillus bacteriocins. FEMS Microbiol. Rev. 2011, 35, 201–232. [Google Scholar] [CrossRef] [Green Version]
- Zendo, T. Screening and characterization of novel bacteriocins from lactic acid bacteria. Biosci. Biotechnol. Biochem. 2013, 77, 893–899. [Google Scholar] [CrossRef]
- Levetin, E.; Dorsey, K. Contribution of leaf surface fungi to the air spora. Aerobiologia 2006, 22, 3–12. [Google Scholar] [CrossRef]
- Pitcher, D.G.; Saunders, N.A.; Owen, R.J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 1989, 8, 151–156. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger dataset. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Bonev, B.; Hooper, J.; Parisot, J. Principles of assessing bacterial susceptibility to antibiotics using the agar diffusion method. J. Antimicrob. Chemo. 2008, 61, 1295–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clinical Laboratory Standards Institute. Performance standards of antimicrobial disc susceptibility tests. CLSI 2012, 32, 3. [Google Scholar]
- Huang, Y.; Adams, M.C. In vitro assessment of the upper gastrointestinal tolerance of potential probiotic dairy propionibacteria. Int. J. Food Microbiol. 2004, 91, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Valeriano, V.D.; Parungao-Balolong, M.M.; Kang, D.K. In vitro evaluation of the mucin-adhesion ability and probiotic potential of Lactobacillus mucosae LM1. J. Appl. Microbiol. 2014, 117, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Llanco, L.A.; Nakano, V.; de Moraes, C.T.P.; Piazza, R.M.F.; Avila-Campos, M.J. Adhesion and invasion of Clostridium perfringens type A into epithelial cells. Braz. J. Microbiol. 2017, 48, 764–768. [Google Scholar] [CrossRef] [PubMed]
- Spellerberg, I.F.; Fedor, P.J. A tribute to Claude Shannon (1916-2001) and a plea for more rigorous use of species richness, species diversity and the ‘Shannon-Wiener’Index. Glob. Ecol. Biogeogr. 2003, 12, 177–179. [Google Scholar] [CrossRef] [Green Version]
- Pielou, E.C. The measurement of diversity in different types of biological collections. J. Theor. Biol. 1966, 13, 131–144. [Google Scholar] [CrossRef]
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Colwell, R.K. Biodiversity: Concepts, Patterns and Measurement. In The Princeton Guide to Ecology; Simon, A.L., Ed.; Princeton University Press: Princeton, NJ, USA, 2009; pp. 257–263. [Google Scholar]
- Yeh, H.Y.; Line, J.E.; Hinton, A., Jr.; Gao, Y.; Zhuang, H. The effect of rosemary extract and cold plasma treatments on bacterial community diversity in poultry ground meats. Heliyon 2019, 5, e02719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cihan, A.C. Taxonomic classification of Anoxybacillus isolates from geothermal regions in Turkey by 16S rRNA gene sequences and ARDRA, ITS-PCR, Rep-PCR analyses. Pol. J. Microbiol. 2013, 62, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Wenzler, E.; Kamboj, K.; Balada-Llasat, J.M. Severe sepsis secondary to persistent Lysinibacillus sphaericus, Lysinibacillus fusiformis and Paenibacillus amylolyticus bacteremia. Int. J. Infect. Dis. 2015, 35, 93–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagier, J.C.; Ramasamy, D.; Rivet, R.; Raoult, D.; Fournier, P.E. Non contiguous-finished genome sequence and description of Cellulomonas massiliensis sp. nov. Stand. Genomic Sci. 2012, 7, 258–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Nishiyama, M.; Kunito, T.; Senoo, K.; Kawahara, K.; Murakami, K.; Oyaizu, H. High population of Sphingomonas species on plant surface. J. Appl. Microbiol. 2004, 85, 731–736. [Google Scholar] [CrossRef]
- Bafana, A. Diversity and metabolic potential of culturable root-associated bacteria from Origanum vulgare in sub-Himalayan region. World J. Microbiol. Biotechnol. 2013, 29, 63–74. [Google Scholar] [CrossRef]
- Walterson, A.M.; Stavrinides, J. Pantoea: Insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol. Rev. 2015, 39, 968–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Undabarrena, A.; Beltrametti, F.; Claverías, F.P.; González, M.; Moore, E.R.B.; Seeger, M.; Cámara, B. Exploring the diversity and antimicrobial potential of marine Actinobacteria from the Comau Fjord in Northern Patagonia, Chile. Front. Microbiol. 2016, 7, 1135. [Google Scholar] [CrossRef]
- Naureen, Z.; Rehman, N.U.; Hussain, H.; Hussain, J.; Gilani, S.A.; Housni, S.K.A.; Mabood, F.; Khan, A.L.; Farooq, S.; Abbas, G.; et al. Exploring the potentials of Lysinibacillus sphaericus ZA9 for plant growth promotion and biocontrol activities against phytopathogenic fungi. Front. Microbiol. 2017, 8, 1477. [Google Scholar] [CrossRef]
- Hussain, A.; Amna; Kamran, M.A.; Javed, M.T.; Hayet, K.; Farooq, M.A.; Ali, N.; Ali, M.; Manghwar, H.; Jan, F.; et al. Individual and combinatorial application of Kocuria rhizophila and citric acid on phytoextraction of multi-metal contaminated soils by Glycine max L. Environ. Exp. Bot. 2019, 159, 23–33. [Google Scholar] [CrossRef]
- Qessaoui, R.; Bouharroud, R.; Furze, J.N.; El Aalaoui, M.; Akroud, H.; Amarraque, A.; Van Vaerenbergh, J.; Tahzima, R.; Mayad, E.H.; Chebli, B. Applications of new rhizobacteria Pseudomonas isolates in agroecology via fundamental processes complementing plant growth. Sci. Rep. 2019, 9, 12832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uniyal, S.; Paliwal, R.; Verma, M.; Sharma, R.K.; Rai, J.P. Isolation and characterization of fipronil degrading Acinetobacter calcoaceticus and Acinetobacter oleivorans from rhizospheric zone of Zea mays. Bull. Environ. Contam. Toxicol. 2016, 96, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Chebbi, A.; Mhiri, N.; Rezgui, F.; Ammar, N.; Maalej, A.; Sayadi, S.; Chamkha, M. Biodegradation of malodorous thiols by a Brevibacillus sp. strain isolated from a Tunisian phosphate factory. FEMS Microbiol. Lett. 2015, 362, fnv097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panda, A.K.; Bisht, S.S.; DeMondal, S.; Kumar, N.S.; Gurusubramanian, G.; Panigrahi, A.K. Brevibacillus as a biological tool: A short review. Antonie van Leeuwenhoek 2014, 105, 623–639. [Google Scholar] [CrossRef] [PubMed]
- Atrouni, A.A.; Joly-Guillou, M.; Hamze, M.; Kempf, M. Reservoirs of non-baumannii Acinetobacter species. Front. Microbiol. 2016, 7, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fusco, V.; Quero, G.M.; Cho, G.S.; Kabisch, J.; Meske, D.; Neve, H.; Bockelmann, W.; Franz, C.M.A.P. The genus Weissella: Taxonomy, ecology and biotechnological potential. Front. Microbiol. 2015, 6, 155. [Google Scholar] [CrossRef] [Green Version]
- Chuah, L.O.; Yap, K.P.; Shamila-Syuhada, A.K.; Thong, K.L.; Ahmad, R.; Liong, M.T.; Rusul, G. Floricoccus tropicus gen. nov., sp. nov. and Floricoccus penangensis sp. nov. isolated from fresh flowers of durian tree and hibiscus. Int. J. Syst. Evol. Microbiol. 2017, 67, 4979–4985. [Google Scholar] [CrossRef] [Green Version]
- Papadimitriou, K.; Alegría, A.; Bron, P.A.; De Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C.; et al. Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 837–890. [Google Scholar] [CrossRef] [Green Version]
- Harvey, Z.H.; Snider, M.J. Draft genome sequence of the nicotinate-metabolizing soil bacterium Bacillus niacini DSM 2923. Genome Announc. 2014, 2, e01251-14. [Google Scholar] [CrossRef] [Green Version]
- Elshaghabee, F.M.F.; Rokana, N.; Gulhane, R.D.; Sharma, C.; Panwar, H. Bacillus as potential probiotics: Status, concerns, and future perspectives. Front. Microbiol. 2017, 8, 1490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Guan, Y.; Dong, Y.; Zhao, L.; Rong, S.; Chen, W.; Lv, M.; Xu, H.; Gao, X.; Chen, R.; et al. Isolation and evaluation of endophytic Bacillus tequilensis GYLH001 with potential application for biological control of Magnaporthe oryzae. PLoS ONE 2018, 13, e0203505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korobov, V.V.; Zhurenko, E.I.; Zharikova, N.V.; Iasakov, T.R.; Markusheva, T.V. Application of the new degrader strain Bacillus mobilis 34T for soil treatment from 2,4,5-trichlorophenoxyacetic acid. Moscow Univ. Biol. Sci. Bull. 2019, 74, 154–157. [Google Scholar] [CrossRef]
- Saran, A.; Imperato, V.; Fernandez, L.; Gkorezis, P.; D’Haen, J.; Merini, L.J.; Vangronsveld, J.; Thijs, S. Phytostabilization of polluted military soil supported by bioaugmentation with PGP-trace element tolerant bacteria isolated from Helianthus petiolaris. Agronomy 2020, 10, 204. [Google Scholar] [CrossRef] [Green Version]
- Coates, R.; Moran, J.; Horsburgh, M.J. Staphylococci: Colonizers and pathogens of human skin. Future Microbiol. 2014, 9, 75–91. [Google Scholar] [CrossRef]
- Ma, A.P.Y.; Jiang, J.; Tun, H.M.; Mauroo, N.F.; Yuen, C.S.; Leung, F.C.C. Complete genome sequence of Staphylococcus xylosus HKUOPL8, a potential opportunistic pathogen of mammals. Genome Announc. 2014, 2, e00653-14. [Google Scholar] [CrossRef] [Green Version]
- Brawand, S.G.; Cotting, K.; Gómez-Sanz, E.; Collaud, A.; Thomann, A.; Brodard, I.; Rodriguez-Campos, S.; Strauss, C.; Perreten, V. Macrococcus canis sp. nov., a skin bacterium associated with infections in dogs. Int. J. Syst. Evol. Microbiol. 2017, 67, 621–626. [Google Scholar] [CrossRef]
- Lo, S.; Thiam, I.; Fall, B.; Ba-Diallo, A.; Diallo, O.F.; Diagne, R.; Dia, M.L.; Ka, R.; Sarr, A.M.; Sow, A.I. Urinary tract infection with Corynebacterium aurimucosum after urethroplasty stricture of the urethra: A case report. J. Med. Case Rep. 2015, 9, 156. [Google Scholar] [CrossRef] [Green Version]
- Monahan, L.G.; DeMaere, M.Z.; Cummins, M.L.; Djordjevic, S.P.; Chowdhury, P.R.; Darling, A.E. High contiguity genome sequence of a multidrug-resistant hospital isolate of Enterobacter hormaechei. Gut Pathog. 2019, 11, 3. [Google Scholar] [CrossRef]
- Asai, N.; Koizumi, Y.; Yamada, A.; Sakanashi, D.; Watanabe, H.; Kato, H.; Shiota, A.; Hagihara, M.; Suematsu, H.; Yamagishi, Y.; et al. Pantoea dispersa bacteremia in an immunocompetent patient: A case report and review of the literature. J. Med. Case Rep. 2019, 13, 33. [Google Scholar] [CrossRef] [Green Version]
- Polyakova, O.; Billor, N. Impact of deciduous tree species on litterfall quality, decomposition rates and nutrient circulation in pine stands. For. Ecol. Manag. 2007, 253, 11–18. [Google Scholar] [CrossRef]
- Xiao, M.; Sun, S.S.; Zhang, Z.Z.; Wang, J.M.; Qiu, L.W.; Sun, H.Y.; Song, Z.Z.; Zhang, B.Y.; Gao, D.L.; Zhang, G.Q.; et al. Analysis of bacterial diversity in two oil blocks from two low-permeability reservoirs with high salinities. Sci. Rep. 2016, 6, 19600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siles, J.A.; Cajthaml, T.; Minerbi, S.; Margesin, R. Effect of altitude and season on microbial activity, abundance and community structure in Alpine forest soils. FEMS Microbiol. Ecol. 2016, 92, fiw008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malele, I.; Nyingilili, H.; Lyaruu, E.; Tauzin, M.; Ollivier, B.B.; Cayol, J.L.; Fardeau, M.L.; Geiger, A. Bacterial diversity obtained by culturable approaches in the gut of Glossina pallidipes population from a non sleeping sickness focus in Tanzania: Preliminary results. BMC Microbiol. 2018, 18, 164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeżewska-Frąckowiak, J.; Żebrowska, J.; Czajkowska, E.; Jasińska, J.; Pęksa, M.; Jędrzejczak, G.; Skowron, P.M. Identification of bacterial species in probiotic consortiums in selected commercial cleaning preparations. Acta Biochim. Pol. 2019, 66, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Unban, K.; Kochasee, P.; Shetty, K.; Khanongnuch, C. Tannin-tolerant and extracellular tannase producing Bacillus isolated from traditional fermented tea leaves and their probiotic functional properties. Foods 2020, 9, 490. [Google Scholar] [CrossRef]
- Amin, F.A.Z.; Sabri, S.; Ismail, M.; Chan, K.W.; Ismail, N.; Esa, N.M.; Lila, M.A.M.; Zawawi, N. Probiotic properties of Bacillus strains isolated from stingless bee (Heterotrigona itama) honey collected across Malaysia. Int. J. Environ. Res. Public Health 2020, 17, 278. [Google Scholar] [CrossRef] [Green Version]
- Kristoffersen, S.M.; Ravnum, S.; Tourasse, N.J.; Økstad, O.A.; Kolstø, A.; Davies, W. Low concentrations of bile salts induce stress responses and reduce motility in Bacillus cereus ATCC 14570. J. Bacteriol. 2007, 189, 5302–5313. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Province | H’ | D | J’ | R |
|---|---|---|---|---|
| Chiang Mai | 0.90 | 0.53 | 0.69 | 26.31 |
| Chiang Rai | 1.20 | 0.53 | 0.56 | 58.39 |
| Lampang | 1.39 | 0.67 | 0.63 | 69.84 |
| Phayao | 1.49 | 0.71 | 0.77 | 46.74 |
| Phrae | 0.89 | 0.53 | 0.73 | 18.73 |
| Nan | 1.01 | 0.51 | 0.56 | 32.30 |
| Bacillus Strain | Zone of Inhibition (mm) | |||
|---|---|---|---|---|
| B. cereus TISTR 687 | E. coli O157:H7 DMST 12743 | S. aureus ATCC 25923 | MRSA DMST 20625 | |
| B. clausii ML062-2 | 0 e | 7.2 ± 0.1 b | 0 d | 0 d |
| B. subtilis ML066-3 | 7.3 ± 0.3 d | 0 c | 9.0 ± 0.0 b | 0 d |
| B. licheniformis ML071-1 | 9.3 ± 0.4 c | 0 c | 0 d | 0 d |
| B. licheniformis ML073-1 | 9.8 ± 0.4 c | 0 c | 0 d | 0 d |
| B. licheniformis ML075-1 | 11.3 ± 0.4 b | 0 c | 9.0 ± 0.0 b | 0 d |
| B. licheniformis ML076-2 | 11.0 ± 0.0 b | 0 c | 0 d | 0 d |
| B. siamensis ML122-2 | 0 e | 0 c | 8.0 ± 0.0 c | 12.0 ± 0.0 a |
| B. siamensis ML123-1 | 0 e | 0 c | 0 d | 9.3 ± 0.4 c |
| B. siamensis ML124-1 | 0 e | 0 c | 0 d | 11.3 ± 0.4 b |
| Gentamicin | 16.0 ± 0.8 a | 11.6 ± 0.4 a | 14.9 ± 0.2 a | 12.1 ± 0.2 a |
| Bacterial Strain | Adhesion Ability (%) | |
|---|---|---|
| L. acidophilus TISTR 2365 | L. plantarum FM03-1 | |
| L. acidophilus TISTR 2365 | 100 b | Not applied |
| L. plantarum FM03-1 | Not applied | 100 b |
| B. licheniformis ML075-1 | 15.4 ± 3.3 c | 52.6 ± 1.6 d |
| B. siamensis ML122-2 | 19.5 ± 1.9 c | 75.8 ± 7.4 c |
| E. coli ATCC 25922 | 127.2 ± 8.2 a | 493.6 ± 31.8 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rungsirivanich, P.; Supandee, W.; Futui, W.; Chumsai-Na-Ayudhya, V.; Yodsombat, C.; Thongwai, N. Culturable Bacterial Community on Leaves of Assam Tea (Camellia sinensis var. assamica) in Thailand and Human Probiotic Potential of Isolated Bacillus spp. Microorganisms 2020, 8, 1585. https://doi.org/10.3390/microorganisms8101585
Rungsirivanich P, Supandee W, Futui W, Chumsai-Na-Ayudhya V, Yodsombat C, Thongwai N. Culturable Bacterial Community on Leaves of Assam Tea (Camellia sinensis var. assamica) in Thailand and Human Probiotic Potential of Isolated Bacillus spp. Microorganisms. 2020; 8(10):1585. https://doi.org/10.3390/microorganisms8101585
Chicago/Turabian StyleRungsirivanich, Patthanasak, Witsanu Supandee, Wirapong Futui, Vipanee Chumsai-Na-Ayudhya, Chaowarin Yodsombat, and Narumol Thongwai. 2020. "Culturable Bacterial Community on Leaves of Assam Tea (Camellia sinensis var. assamica) in Thailand and Human Probiotic Potential of Isolated Bacillus spp." Microorganisms 8, no. 10: 1585. https://doi.org/10.3390/microorganisms8101585