First Isolation and Characterization of Bacteria from the Core’s Cooling Pool of an Operating Nuclear Reactor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Bacterial Strains
2.2. Identification of Bacterial Strains
2.3. Characterization of Survival to Irradiation
2.4. Microorganism Exposure to Uranium
2.5. Uranium Quantitation by Mass Spectrometry
3. Results and Discussion
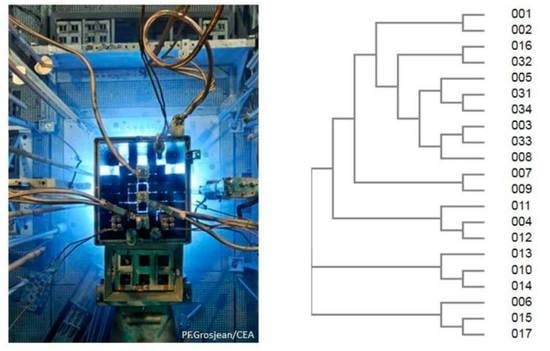
3.1. Identifying Microorganisms in the Reactor Pool after Cultivation
3.2. Assessment of Radiation Tolerance
3.3. Uptake of Uranium
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vandenhove, H. European sites contaminated by residues from the ore-extracting and -processing industries. Int. Congr. Ser. 2002, 1225, 307–315. [Google Scholar] [CrossRef]
- Wetterlind, J.; De Forges, A.C.R.; Nicoullaud, B.; Arrouays, D. Changes in uranium and thorium contents in topsoil after long-term phosphorus fertilizer application. Soil Use Manag. 2012, 28, 101–107. [Google Scholar] [CrossRef]
- Yamazaki, I.M.; Geraldo, L.P. Uranium content in phosphate fertilizers commercially produced in Brazil. Appl. Radiat. Isot. 2003, 59, 133–136. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Huang, Z.; Liu, H.; Hou, J.; Liu, X. Advances on the toxicity of uranium to different organisms. Chemosphere 2019, 237, 124548. [Google Scholar] [CrossRef] [PubMed]
- Anke, M.; Seeber, O.; Müller, R.; Schäfer, U.; Zerull, J. Uranium transfer in the food chain from soil to plants, animals and man. Geochemistry 2009, 69, 75–90. [Google Scholar] [CrossRef]
- Efremenkov, V.M. Radioactive waste management at nuclear power plants. IAEA Bull. 1989, 31, 37. [Google Scholar]
- International Atomic Energy Agency. Combined Methods for Liquid Radioactive Waste Treatment; International Atomic Energy Agency: Vienna, Austria, 2003. [Google Scholar]
- Godheja, J.; Shekhar, S.; Siddiqui, S.; Modi, D. Xenobiotic Compounds Present in Soil and Water: A Review on Remediation Strategies. J. Environ. Anal. Toxicol. 2016, 6, 392. [Google Scholar] [CrossRef]
- Lloyd, J.R.; Renshaw, J.C. Bioremediation of radioactive waste: Radionuclide–microbe interactions in laboratory and field-scale studies. Curr. Opin. Biotechnol. 2005, 16, 254. [Google Scholar] [CrossRef]
- Newsome, L.; Morris, K.; Lloyd, J.R. The biogeochemistry and bioremediation of uranium and other priority radionuclides. Chem. Geol. 2014, 363, 164–184. [Google Scholar] [CrossRef]
- Galã¨s, G.; Libert, M.-F.; Sellier, R.; Cournac, L.; Chapon, V.; Heulin, T. Molecular hydrogen from water radiolysis as an energy source for bacterial growth in a basin containing irradiating waste. FEMS Microbiol. Lett. 2004, 240, 155–162. [Google Scholar] [CrossRef] [Green Version]
- Chicote, E.; García, A.M.; Moreno, D.A.; Sarró, M.I.; Lorenzo, P.I.; Montero, F. Isolation and identification of bacteria from spent nuclear fuel pools. J. Ind. Microbiol. Biotechnol. 2005, 32, 155–162. [Google Scholar] [CrossRef]
- Sarró, M.I.; Garcia, A.M.; Moreno, D.A. Biofilm formation in spent nuclear fuel pools and bioremediation of radioactive water. Int. Microbiol. 2005, 8, 223–230. [Google Scholar] [PubMed]
- Masurat, P.; Fru, E.; Pedersen, K. Identification of Meiothermus as the dominant genus in a storage system for spent nuclear fuel. J. Appl. Microbiol. 2005, 98, 727–740. [Google Scholar] [CrossRef] [PubMed]
- Foster, L.; Boothman, C.; Ruiz-Lopez, S.; Boshoff, G.; Jenkinson, P.; Sigee, D.; Pittman, J.K.; Morris, K.; Lloyd, J.R. Microbial bloom formation in a high pH spent nuclear fuel pond. Sci. Total. Environ. 2020, 720, 137515. [Google Scholar] [CrossRef]
- Rivasseau, C.; Farhi, E.; Compagnon, E.; Cyr, D.D.G.S.; van Lis, R.; Falconet, D.; Kuntz, M.; Atteia, A.; Couté, A. Coccomyxa actinabiotis sp. nov. (Trebouxiophyceae, Chlorophyta), a new green microalga living in the spent fuel cooling pool of a nuclear reactor. J. Phycol. 2016, 52, 689–703. [Google Scholar] [CrossRef]
- MeGraw, V.E.; Brown, A.R.; Boothman, C.; Goodacre, R.; Morris, K.; Sigee, D.; Anderson, L.; Lloyd, J.R. A Novel Adaptation Mechanism Underpinning Algal Colonization of a Nuclear Fuel Storage Pond. mBio 2018, 9, e02395-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chicote, E.; Moreno, D.A.; Garcia, A.M.; Sarro, M.I.; Lorenzo, P.I.; Montero, F. Biofouling on the Walls of a Spent Nuclear Fuel Pool with Radioactive Ultrapure Water. Biofouling 2004, 20, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Silva, R.; de Almeida, D.M.; Cabral, B.C.A.; Dias, V.H.G.; Mello, I.C.D.T.E.; Ürményi, T.P.; Woerner, A.E.; Neto, R.S.D.M.; Budowle, B.; Nassar, C.A.G. Microbial enrichment and gene functional categories revealed on the walls of a spent fuel pool of a nuclear power plant. PLoS ONE 2018, 13, e0205228. [Google Scholar] [CrossRef] [Green Version]
- Petit, P.C.M.; Pible, O.; Eesbeeck, V.V.; Alban, C.; Steinmetz, G.; Mysara, M.; Monsieurs, P.; Armengaud, J.; Rivasseau, C. Direct Meta-Analyses Reveal Unexpected Microbial Life in the Highly Radioactive Water of an Operating Nuclear Reactor Core. Microorganisms 2020, 8, 1857. [Google Scholar] [CrossRef]
- Ruiz-Lopez, S.; Foster, L.; Boothman, C.; Cole, N.; Morris, K.; Lloyd, J.R. Identification of a Stable Hydrogen-Driven Microbiome in a Highly Radioactive Storage Facility on the Sellafield Site. Front. Microbiol. 2020, 11, 587556. [Google Scholar] [CrossRef]
- Bagwell, C.E.; Noble, P.A.; Milliken, C.E.; Li, D.; Kaplan, D.I. Amplicon Sequencing Reveals Microbiological Signatures in Spent Nuclear Fuel Storage Basins. Front. Microbiol. 2018, 9, 377. [Google Scholar] [CrossRef] [Green Version]
- Bruhn, D.F.; Breckenridge, C.R.; Tsang, M.N.; Watkins, C.S.; Windes, W.E.; Roberto, F.F.; Tsang, M.N.; Pinhero, P.J.; Brey, R.F.; Wright, R.N.; et al. Irradiation of Microbes from Spent Nuclear Fuel Storage Pool Environments; Global 99, ANS: Jackson Hole, WY, USA, 1999. [Google Scholar]
- Tišáková, L.; Pipíška, M.; Godány, A.; Horník, M.; Vidová, B.; Augustín, J. Bioaccumulation of 137Cs and 60Co by bacteria isolated from spent nuclear fuel pools. J. Radioanal. Nucl. Chem. 2013, 295, 737–748. [Google Scholar] [CrossRef]
- Rivasseau, C.; Farhi, E.; Atteia, A.; Couté, A.; Gromova, M.; Cyr, D.d.G.S.; Boisson, A.-M.; Féret, A.-S.; Compagnon, E.; Bligny, R. An extremely radioresistant green eukaryote for radionuclide bio-decontamination in the nuclear industry. Energy Environ. Sci. 2013, 6, 1230–1239. [Google Scholar] [CrossRef] [Green Version]
- Gerber, U.; Hübner, R.; Rossberg, A.; Krawczyk-Bärsch, E.; Merroun, M.L. Metabolism-dependent bioaccumulation of uranium by Rhodosporidium toruloides isolated from the flooding water of a former uranium mine. PLoS ONE 2018, 13, e0201903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slade, D.; Radman, M. Oxidative Stress Resistance in Deinococcus radiodurans. Microbiol. Mol. Biol. Rev. 2011, 75, 133–191. [Google Scholar] [CrossRef] [Green Version]
- Van Eesbeeck, V.; Props, R.; Mysara, M.; Petit, P.C.M.; Rivasseau, C.; Armengaud, J.; Monsieurs, P.; Mahillon, J.; Leys, N. Cyclical Patterns Affect Microbial Dynamics in the Water Basin of a Nuclear Research Reactor. Front. Microbiol. 2021, 12, 744115. [Google Scholar] [CrossRef]
- Pible, O.; Allain, F.; Jouffret, V.; Culotta, K.; Miotello, G.; Armengaud, J. Estimating relative biomasses of organisms in microbiota using “phylopeptidomics”. Microbiome 2020, 8, 30. [Google Scholar] [CrossRef] [Green Version]
- Armengaud, J. Metaproteomics to understand how microbiota function: The crystal ball predicts a promising future. Environ. Microbiol. 2023, 25, 115–125. [Google Scholar] [CrossRef]
- Pible, O.; Petit, P.; Steinmetz, G.; Rivasseau, C.; Armengaud, J. Taxonomical composition and functional analysis of biofilms sampled from a nuclear storage pool. Front. Microbiol. 2023, 14, 1148976. [Google Scholar] [CrossRef]
- Hayoun, K.; Pible, O.; Petit, P.; Allain, F.; Jouffret, V.; Culotta, K.; Rivasseau, C.; Armengaud, J.; Alpha-Bazin, B. Proteotyping Environmental Microorganisms by Phylopeptidomics: Case Study Screening Water from a Radioactive Material Storage Pool. Microorganisms 2020, 8, 1525. [Google Scholar] [CrossRef]
- Giacobone, A.F.; Rodriguez, S.A.; Burkart, A.L.; Pizarro, R.A. Microbiological induced corrosion of AA 6061 nuclear alloy in highly diluted media by Bacillus cereus RE 10. Int. Biodeterior. Biodegrad. 2011, 65, 1161–1168. [Google Scholar] [CrossRef]
- Sarró, M.I.; Moreno, D.A.; Chicote, E.; Lorenzo, P.I.; García, A.M.; Montero, F. Biofouling on austenitic stainless steels in spent nuclear fuel pools. Mater. Corros. 2003, 54, 535–540. [Google Scholar] [CrossRef]
- Ruiz-González, M.X.; Czirják, G.; Genevaux, P.; Møller, A.P.; Mousseau, T.A.; Heeb, P. Resistance of Feather-Associated Bacteria to Intermediate Levels of Ionizing Radiation near Chernobyl. Sci. Rep. 2016, 6, 22969. [Google Scholar] [CrossRef]
- Bruhn, D.; Frank, S.; Roberto, F.; Pinhero, P.; Johnson, S. Microbial biofilm growth on irradiated, spent nuclear fuel cladding. J. Nucl. Mater. 2009, 384, 140–145. [Google Scholar] [CrossRef]
- Hu, X.; Mallikarjunan, P.; Koo, J.; Andrews, L.S.; Jahncke, M.L. Comparison of Kinetic Models to Describe High Pressure and Gamma Irradiation Used To Inactivate Vibrio vulnificus and Vibrio parahaemolyticus Prepared in Buffer Solution and in Whole Oysters. J. Food Prot. 2005, 68, 292–295. [Google Scholar] [CrossRef] [Green Version]
- Farag, M.D.E.-D.H.; Shamsuzzaman, K.; Borsa, J. Radiation Sensitivity of Listeria monocytogenes in Phosphate Buffer, Trypticase Soy Broth, and Poultry Feed. J. Food Prot. 1990, 53, 648–651. [Google Scholar] [CrossRef] [PubMed]
- Gursel, B.; Gurakan, G. Effects of gamma irradiation on the survival of Listeria monocytogenes and on its growth at refrigeration temperature in poultry and red meat. Poult. Sci. 1997, 76, 1661–1664. [Google Scholar] [CrossRef] [PubMed]
- Shukla, M.; Chaturvedi, R.; Tamhane, D.; Vyas, P.; Archana, G.; Apte, S.; Bandekar, J.; Desai, A. Multiple-Stress Tolerance of Ionizing Radiation-Resistant Bacterial Isolates Obtained from Various Habitats: Correlation Between Stresses. Curr. Microbiol. 2007, 54, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Ordoñez, O.F.; Flores, M.R.; Dib, J.R.; Paz, A.; Farías, M.E. Extremophile Culture Collection from Andean Lakes: Extreme Pristine Environments that Host a Wide Diversity of Microorganisms with Tolerance to UV Radiation. Microb. Ecol. 2009, 58, 461. [Google Scholar] [CrossRef] [PubMed]
- Briggs, A. The Resistances of Spores of the Genus Bacillus to Phenol, Heat and Radiation. J. Appl. Bacteriol. 1966, 29, 490. [Google Scholar] [CrossRef]
- Setlow, P. Spores of Bacillus subtilis: Their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 2006, 101, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Dussault, D.; Caillet, S.; Le Tien, C.; Lacroix, M. Carotenoids’ influence on radiotolerance of Pantoea agglomerans, a plant pathogen. Lett. Appl. Microbiol. 2008, 47, 208. [Google Scholar] [CrossRef] [PubMed]
- Rainey, F.A.; Ray, K.; Ferreira, M.; Gatz, B.Z.; Nobre, M.F.; Bagaley, D.; Rash, B.A.; Park, M.J.; Earl, A.M. Extensive Diversity of Ionizing-Radiation-Resistant Bacteria Recovered from Sonoran Desert Soil and Description of Nine New Species of the Genus Deinococcus Obtained from a Single Soil Sample. Appl. Environ. Microbiol. 2005, 71, 5225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrie, L.; North, N.N.; Dollhopf, S.L.; Balkwill, D.L.; Kostka, J.E. Enumeration and characterization of iron(III)-reducing microbial communities from acidic subsurface sediments contaminated with uranium(VI). Appl. Environ. Microbiol. 2003, 69, 7467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panak, P.; Raff, J.; Selenska-Pobell, S.; Geipel, G.; Bernhard, G.; Nitsche, H. Complex formation of U(VI) with Bacillus-isolates from a uranium mining waste pile. Radiochim. Acta 2000, 88, 71–76. [Google Scholar] [CrossRef]
- Li, X.; Ding, C.; Liao, J.; Lan, T.; Li, F.; Zhang, D.; Yang, J.; Yang, Y.; Luo, S.; Tang, J.; et al. Biosorption of uranium on Bacillus sp. dwc-2: Preliminary investigation on mechanism. J. Environ. Radioact. 2014, 135, 6–12. [Google Scholar] [CrossRef]
- Nakajima, A.; Tsuruta, T. Competitive biosorption of thorium and uranium by Micrococcus luteus. J. Radioanal. Nucl. Chem. 2004, 260, 13–18. [Google Scholar] [CrossRef]
- Hennig, C.; Panak, P.J.; Reich, T.; Roßberg, A.; Raff, J.; Selenska-Pobell, S.; Matz, W.; Bucher, J.J.; Bernhard, G.; Nitsche, H. EXAFS investigation of uranium(VI) complexes formed at Bacillus cereus and Bacillus sphaericus surfaces. Radiochim. Acta 2001, 89, 625–632. [Google Scholar] [CrossRef]
- Banala, U.K.; Das, N.P.I.; Toleti, S.R. Uranium sequestration abilities of Bacillus bacterium isolated from an alkaline mining region. J. Hazard. Mater. 2021, 411, 125053. [Google Scholar] [CrossRef]
- Fowle, D.A.; Fein, J.B.; Martin, A.M. Experimental Study of Uranyl Adsorption onto Bacillus subtilis. Environ. Sci. Technol. 2000, 34, 3737–3741. [Google Scholar] [CrossRef]
- Ozdemir, S.; Oduncu, M.K.; Kilinc, E.; Soylak, M. Resistance, bioaccumulation and solid phase extraction of uranium (VI) by Bacillus vallismortis and its UV–vis spectrophotometric determination. J. Environ. Radioact. 2017, 171, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Nazina, T.N.; Luk’yanova, E.A.; Zakharova, E.V.; Ivoĭlov, V.S.; Poltaraus, A.B.; Kalmykov, S.N.; Belyaev, S.S.; Zubkov, A.A. Distribution and activity of microorganisms in the deep repository for liquid radioactive waste at the Siberian Chemical Combine. Mikrobiologiia 2006, 75, 836. [Google Scholar] [CrossRef] [PubMed]
- Nazina, T.N.; Luk’Yanova, E.A.; Zakharova, E.V.; Konstantinova, L.I.; Kalmykov, S.N.; Poltaraus, A.B.; Zubkov, A.A. Microorganisms in a Disposal Site for Liquid Radioactive Wastes and Their Influence on Radionuclides. Geomicrobiol. J. 2010, 27, 473–486. [Google Scholar] [CrossRef]
- Tsuruta, T. Removal and recovery of uranyl ion using various microorganisms. J. Biosci. Bioeng. 2002, 94, 23. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, H.; Song, W.; Ma, W.; Hu, W.; Chen, T.; Liu, L. Accumulation of U(VI) on the Pantoea sp. TW18 isolated from radionuclide-contaminated soils. J. Environ. Radioact. 2018, 192, 219. [Google Scholar] [CrossRef]
- Ding, L.; Tan, W.-F.; Xie, S.-B.; Mumford, K.; Lv, J.-W.; Wang, H.-Q.; Fang, Q.; Zhang, X.-W.; Wu, X.-Y.; Li, M. Uranium adsorption and subsequent re-oxidation under aerobic conditions by Leifsonia sp.—Coated biochar as green trapping agent. Environ. Pollut. 2018, 242, 778–787. [Google Scholar] [CrossRef]
- Goodfellow, M.; Kämpfer, P.; Busse, H., Jr.; Trujillo, M.E.; Suzuki, K.-i.; Ludwig, W.; Suzuki, K.I.; Parte, A. Bergey’s Manual of Systematic Bacteriology; Volume 5 the Actinobacteria. Part B; Springer: New York, NY, USA, 2012. [Google Scholar]
- Tsukamura, M. Proposal of a New Genus, Gordona, for Slightly Acid-fast Organisms Occurring in Sputa of Patients with Pulmonary Disease and in Soil. J. Gen. Microbiol. 1971, 68, 15–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Sampling | Phylum/Class | Species | Strain |
|---|---|---|---|
| Reactor operating, 2015 campaign | Actinobacteria | Gordonia bronchialis | CEA-012 |
| Kocuria koreensis | CEA-006 | ||
| Leifsonia sp. | CEA-010 | ||
| CEA-013 CEA-014 | |||
| Micrococcus luteus | CEA-001 CEA-002 | ||
| Rhodococcus corynebacterioides | CEA-004 | ||
| Rothia mucilaginosa | CEA-015 | ||
| CEA-017 | |||
| Streptomyces canus | CEA-011 | ||
| Firmicutes | Bacillus altitudinis | CEA-003 | |
| CEA-008 | |||
| CEA-033 | |||
| Bacillus thuringiensis | CEA-031 | ||
| CEA-034 | |||
| Brevibacillus agri | CEA-032 | ||
| Staphylococcus epidermidis | CEA-005 | ||
| Streptococcus sanguinis | CEA-016 | ||
| α-Proteobacteria | Rhizobiales sp. | CEA-007 | |
| γ-Proteobacteria | Acinetobacter johnsonii | CEA-009 | |
| Reactor shutdown, 2017 campaign | Actinobacteria | Cellulomonas sp. | CEA-434 CEA-444 |
| Mycobacterium aubagnense | CEA-021 | ||
| Mycobacterium sp. | CEA-022 | ||
| Nocardia niigatensis | CEA-020 CEA-030 | ||
| Firmicutes | Bacillus sp. | CEA-448 | |
| α-Proteobacteria | Afipia sp. | CEA-026 | |
| Bradyrhizobium sp. | CEA-023 CEA-025 | ||
| Sphingomonas echinoides | CEA-027 CEA-028 | ||
| Sphingomonas sp. | CEA-154 CEA-87 | ||
| β-Proteobacteria | Pelomonas puraquae | CEA-019 CEA-024 | |
| Pelomonas sp. | CEA-018 CEA-104 CEA-111 | ||
| Ralstonia pickettii | CEA-029 CEA-163 | ||
| γ-Proteobacteria | Pantoea vagans | CEA-450 | |
| Pantoea sp. | CEA-497 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petit, P.; Hayoun, K.; Alpha-Bazin, B.; Armengaud, J.; Rivasseau, C. First Isolation and Characterization of Bacteria from the Core’s Cooling Pool of an Operating Nuclear Reactor. Microorganisms 2023, 11, 1871. https://doi.org/10.3390/microorganisms11081871
Petit P, Hayoun K, Alpha-Bazin B, Armengaud J, Rivasseau C. First Isolation and Characterization of Bacteria from the Core’s Cooling Pool of an Operating Nuclear Reactor. Microorganisms. 2023; 11(8):1871. https://doi.org/10.3390/microorganisms11081871
Chicago/Turabian StylePetit, Pauline, Karim Hayoun, Béatrice Alpha-Bazin, Jean Armengaud, and Corinne Rivasseau. 2023. "First Isolation and Characterization of Bacteria from the Core’s Cooling Pool of an Operating Nuclear Reactor" Microorganisms 11, no. 8: 1871. https://doi.org/10.3390/microorganisms11081871