Expression of Scytonemin Biosynthesis Genes under Alternative Stress Conditions in the Cyanobacterium Nostoc punctiforme
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain, Culture Conditions, and Pigment Extraction
2.2. Expression of Scytonemin-Associated Genes
3. Results
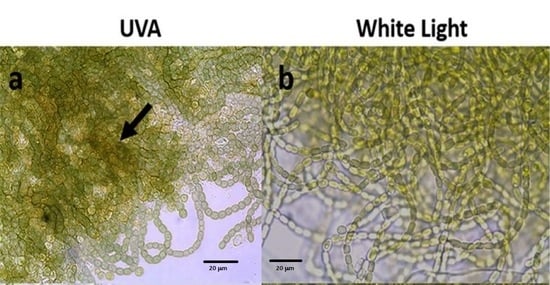
3.1. Scytonemin Production
3.2. Gene Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Castenholz, R.W.; Garcia-Pichel, F. Cyanobacterial responses to UV radiation. In Ecology of Cyanobacteria II; Whitton, B.A., Ed.; Springer: Dordrecht, The Netherlands, 2012; pp. 481–502. [Google Scholar]
- Jagger, J. Solar-UV Actions on Living Cells; Praeger: New York, NY, USA, 1985. [Google Scholar]
- Kasting, J.F. The Proterozoic Biosphere: A Multidisciplinary Study; Cambridge University Press: Cambridge, UK, 1992. [Google Scholar]
- Garcia-Pichel, F. Solar ultraviolet and the evolutionary history of cyanobacteria. Orig. Life Evol. Biosph. 1998, 28, 321–347. [Google Scholar] [CrossRef]
- Jiang, Y.; Rabbi, M.; Kim, M.; Ke, C.; Lee, W.; Clark, R.L.; Mieczkowski, P.A.; Marszalek, P.E. UVA generates pyrimidine dimers in DNA directly. Biophys. J. 2009, 96, 1151–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Baalen, C. The effects of ultraviolet radiation on a coccoid blue-green alga: Survival, photosynthesis, and photoreactivation. Plant Physiol. 1968, 43, 1689–1695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donkor, V.A.; Häder, D.P. Effects of ultraviolet irradiation on photosynthetic pigments in some filamentous cyanobacteria. Aquat. Microbial. Ecol. 1996, 11, 143–149. [Google Scholar] [CrossRef] [Green Version]
- Tyrell, R.M. UVA (320–380 nm) radiation as an oxidative stress. In Oxidative Stress: Oxidants and Antioxidants; Sies, H., Ed.; Academic Press: Berkeley, CA, USA, 1991; pp. 57–83. [Google Scholar]
- Ehling-Schulz, M.; Scherer, S. UV protection in cyanobacteria. Eur. J. Phycol. 1999, 34, 329–338. [Google Scholar] [CrossRef]
- Bebout, B.M.; Garcia-Pichel, F. UV-B-induced vertical migrations of cyanobacteria in a microbial mat. Appl. Environ. Microbiol. 1995, 61, 4215–4222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canfield, L.M.; Forage, J.W.; Valenzuela, J.G. Carotenoids as cellular antioxidants. Proc. Soc. Exp. Biol. Med. 1992, 200, 260–265. [Google Scholar] [CrossRef]
- Garcia-Pichel, F.; Castenholz, R.W. Characterization and biological implications of scytonemin, a cyanobacterial sheath pigment. J. Phycol. 1991, 27, 395–409. [Google Scholar] [CrossRef]
- Garcia-Pichel, F.; Sherry, N.D.; Castenholz, R.W. Evidence for an ultraviolet sunscreen role of the extracellular pigment scytonemin in the terrestrial cyanobacterium Chlorogloeopsis sp. Photochem. Photobiol. 1992, 56, 17–23. [Google Scholar] [CrossRef]
- Matsui, K.; Nazifi, E.; Hirai, Y.; Wada, N.; Matsugo, S.; Sakamoto, T. The cyanobacterial UV-absorbing pigment scytonemin displays radical-scavenging activity. J. Gen. Appl. Microbiol. 2012, 58, 137–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevenson, C.S.; Capper, E.A.; Roshak, A.K.; Marquez, B.; Eichman, C.; Jackson, J.R.; Mattern, M.; Gerwick, W.H.; Jacobs, R.S.; Marshall, L.A. The identification and characterization of the marine natural product scytonemin as a novel antiproliferative pharmacophore. J. Pharmacol. Exp. Ther. 2002, 303, 858–866. [Google Scholar] [CrossRef]
- Duan, Z.F.; Ji, D.N.; Weinstein, E.J.; Liu, X.Z.; Susa, M.; Choy, E.; Yang, C.; Mankin, H.; Hornicek, F.J. Lentiviral shRNA screen of human kinases identifies PLK1 as a potential therapeutic target for osteosarcoma. Cancer Lett. 2010, 293, 220–229. [Google Scholar] [CrossRef]
- Evans, J.; Jones, A.C.; Blumenthal, E.; Soule, T. Anti-proliferation of melanoma cells and immune stimulation by the cyanobacterial indole-alkaloid scytonemin. Fine Focus 2021, 7, 54–63. [Google Scholar] [CrossRef]
- Soule, T.; Palmer, K.; Gao, Q.; Potrafka, R.; Stout, V.; Garcia-Pichel, F. A comparative genomics approach to understanding the biosynthesis of the sunscreen scytonemin in cyanobacteria. BMC Gen. 2009, 10, 336–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soule, T.; Stout, V.; Swingley, W.D.; Meeks, J.C.; Garcia-Pichel, F. Molecular genetics and genomic analysis of scytonemin biosynthesis in Nostoc punctiforme ATCC 29133. J. Bacteriol. 2007, 189, 4465–4472. [Google Scholar] [CrossRef] [Green Version]
- Naurin, S.; Bennett, J.; Videau, P.; Philmus, B.; Soule, T. The response regulator Npun_F1278 is essential for scytonemin biosynthesis in the cyanobacterium Nostoc punctiforme ATCC 29133. J. Phycol. 2016, 52, 564. [Google Scholar] [CrossRef]
- Garcia-Pichel, F.; Lombard, J.; Soule, T.; Dunaj, S.; Wu, S.H.; Wojciechowski, M.F.; Giovannoni, S.J. Timing the evolutionary advent of cyanobacteria and the later great oxidation event using gene phylogenies of a sunscreen. mBio 2019, 10, e00561-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashby, M.; Houmard, J. Cyanobacterial two-component proteins: Structure, diversity, distribution, and evolution. Microbiol. Mol. Biol. Rev. 2006, 70, 472–509. [Google Scholar] [CrossRef] [Green Version]
- Ponting, C.P.; Aravind, L. PAS: A multifunctional domain family comes to light. Curr. Biol. 1997, 7, R674–R677. [Google Scholar] [CrossRef] [Green Version]
- Klicki, K.; Ferreira, D.; Hamill, D.; Dirks, B.; Mitchell, N.; Garcia-Pichel, F.; Greenberg, E.P.; Burnap, R.; Preston, G.; Vermaas, W. The widely conserved ebo cluster is involved in precursor transport to the periplasm during scytonemin synthesis in Nostoc punctiforme. mBio 2018, 9, e02266-18. [Google Scholar] [CrossRef] [Green Version]
- Balskus, E.P.; Walsh, C.T. Investigating the initial steps in the biosynthesis of cyanobacterial sunscreen scytonemin. J. Am. Chem. Soc. 2008, 130, 15260–15261. [Google Scholar] [CrossRef] [Green Version]
- Balskus, E.P.; Walsh, C.T. An enzymatic cyclopentyl[b]indole formation involved in scytonemin biosynthesis. J. Am. Chem. Soc. 2009, 131, 14648–14649. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, D.; Garcia-Pichel, F. Mutational Studies of Putative Biosynthetic Genes for the Cyanobacterial Sunscreen Scytonemin in Nostoc punctiforme ATCC 29133. Front. Microbiol. 2016, 7, 735. [Google Scholar] [CrossRef] [Green Version]
- Dillon, J.G.; Tatsumi, C.M.; Tandingan, P.G.; Castenholz, R.W. Effect of environmental factors on the synthesis of scytonemin, a UV-screening pigment, in a cyanobacterium (Chroococcidiopsis sp.). Arch. Microbiol. 2002, 177, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Fleming, E.D.; Castenholz, R.W. Effects of periodic desiccation on the synthesis of the UV-screening compound, scytonemin, in cyanobacteria. Environ. Microbiol. 2007, 9, 1448–1455. [Google Scholar] [CrossRef] [PubMed]
- Soule, T.; Garcia-Pichel, F.; Stout, V. Gene expression patterns associated with the biosynthesis of the sunscreen scytonemin in Nostoc punctiforme ATCC 29133 in response to UVA radiation. J. Bacteriol. 2009, 191, 4639–4646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janssen, J.; Soule, T. Gene expression of a two-component regulatory system associated with sunscreen biosynthesis in the cyanobacterium Nostoc punctiforme ATCC 29133. FEMS Microbiol. Lett. 2016, 363, fnv235. [Google Scholar] [CrossRef]
- Soule, T.; Gao, Q.; Stout, V.; Garcia-Pichel, F. The global response of Nostoc punctiforme ATCC 29133 to UVA stress, assessed in a temporal DNA microarray study. Photochem. Photobiol. 2013, 89, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Soule, T.; Ferreira, D.; Lothamer, J.; Garcia-Pichel, F. The independent and shared transcriptomic response to UVA, UVB and oxidative stress in the cyanobacterium Nostoc punctiforme ATCC 29133. Photochem. Photobiol. 2021, 97, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.; Arnon, D.I. Studies on nitrogen-fixing blue-green algae. I. Growth and nitrogen-fixation by Anabaena cylindrica. Plant Physiol. 1955, 30, 366–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, E.L.; Summers, M.L.; Christman, H.; Martin, M.E.; Meeks, J.C. Global gene expression patterns of Nostoc punctiforme in steady-state dinitrogen-grown heterocyst-containing cultures at single time points during the differentiation of akinetes and hormogonia. J. Bacteriol. 2007, 186, 5247–5256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorrels, C.M.; Proteau, P.J.; Gerwick, W.H. Organization, evolution, and expression analysis of the biosynthetic gene cluster for scytonemin, a cyanobacterial UV-absorbing pigment. Appl. Environ. Microbiol. 2009, 75, 4861–4869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 2002–2007. [Google Scholar] [CrossRef] [PubMed]


| Stress | Scytonemin | Gene | Fold Change | p-Value |
|---|---|---|---|---|
| UVA | Yes | scyA | +26.05 | 0.049 * |
| trpB | +77.53 | 0.039 * | ||
| eboE | −1.63 | 0.278 | ||
| UVB | Yes | scyA | +22.7 | 0.002 * |
| trpB | +2.23 | 0.001 * | ||
| eboE | −3.01 | 0.001 * | ||
| High Light | No | scyA | +28.59 | <0.001 * |
| trpB | +4.63 | <0.001 * | ||
| eboE | +3.59 | <0.001 * | ||
| Osmotic | No | scyA | −4.90 | 0.002 * |
| trpB | −3.12 | 0.001 * | ||
| eboE | −1.53 | 0.010 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bennett, J.; Soule, T. Expression of Scytonemin Biosynthesis Genes under Alternative Stress Conditions in the Cyanobacterium Nostoc punctiforme. Microorganisms 2022, 10, 427. https://doi.org/10.3390/microorganisms10020427
Bennett J, Soule T. Expression of Scytonemin Biosynthesis Genes under Alternative Stress Conditions in the Cyanobacterium Nostoc punctiforme. Microorganisms. 2022; 10(2):427. https://doi.org/10.3390/microorganisms10020427
Chicago/Turabian StyleBennett, Janine, and Tanya Soule. 2022. "Expression of Scytonemin Biosynthesis Genes under Alternative Stress Conditions in the Cyanobacterium Nostoc punctiforme" Microorganisms 10, no. 2: 427. https://doi.org/10.3390/microorganisms10020427