Identification of a Neisseria gonorrhoeae Histone Deacetylase: Epigenetic Impact on Host Gene Expression
Abstract
:1. Introduction
2. Results
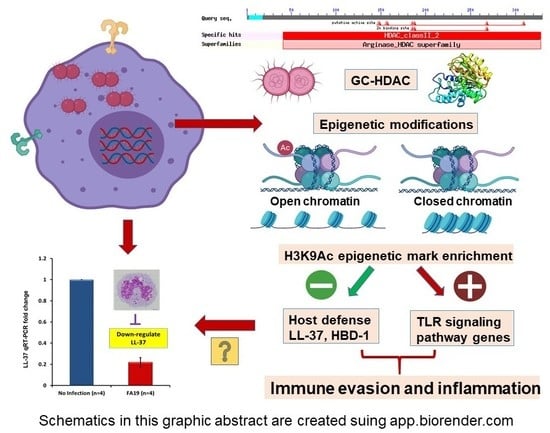
2.1. Gonococcal Infection Downregulates Host Defense Peptides Expression in Macrophages
2.2. Neisseria Gonorrhoeae Contains a Gene Encoding a Histone Deacetylase-Like (Gc-HDAC) Enzyme
2.3. Computational Analysis of Gc-HDAC Enzyme
2.4. Expression of the Gonococcal HDAC-Encoding Gene
2.5. N. Gonorrhoeae Exerts Epigenetic Modifications on Host Innate Immune Genes in Infected Macrophages
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rowley, J.; Vander Hoorn, S.; Korenromp, E.; Low, N.; Unemo, M.; Abu-Raddad, L.J.; Chico, R.M.; Smolak, A.; Newman, L.; Gottlieb, S.; et al. Chlamydia, gonorrhoea, trichomoniasis and syphilis: Global prevalence and incidence estimates, 2016. Bull. World Health Organ. 2019, 97, 548–562. [Google Scholar] [CrossRef]
- CDC. Trends in aging—United States and worldwide. MMWR Morb. Mortal. Wkly. Rep. 2003, 52, 101–104. [Google Scholar]
- Tapsall, J.W.; Shultz, T.; Limnios, E.; Munro, R.; Mercer, J.; Porritt, R.; Griffith, J.; Hogg, G.; Lum, G.; Lawrence, A.; et al. Surveillance of antibiotic resistance in invasive isolates of Neisseria meningitidis in Australia 1994–1999. Pathology 2001, 33, 359–361. [Google Scholar] [CrossRef] [PubMed]
- Shultz, T.R.; Tapsall, J.W.; White, P.A. Correlation of in vitro susceptibilities to newer quinolones of naturally occurring quinolone-resistant Neisseria gonorrhoeae strains with changes in GyrA and ParC. Antimicrob. Agents Chemother. 2001, 45, 734–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. Emergence of Multi-Drug Resistant Neisseria Gonorrhoeae—Threat of Global Rise in Untreated Sexually Transmitted Infections; WHO: Geneva, Switzerland, 2011. [Google Scholar]
- WHO. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Zughaier, S.M.; Kandler, J.L.; Shafer, W.M. Neisseria gonorrhoeae modulates iron-limiting innate immune defenses in macrophages. PLoS ONE 2014, 9, e87688. [Google Scholar] [CrossRef] [PubMed]
- Criss, A.K.; Seifert, H.S. A bacterial siren song: Intimate interactions between Neisseria and neutrophils. Nat. Rev. Microbiol. 2012, 10, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Zughaier, S.M.; Kandler, J.L.; Balthazar, J.T.; Shafer, W.M. Phosphoethanolamine modification of neisseria gonorrhoeae lipid A reduces autophagy flux in macrophages. PLoS ONE 2015, 10, e0144347. [Google Scholar] [CrossRef] [PubMed]
- Kandler, J.L.; Joseph, S.J.; Balthazar, J.T.; Dhulipala, V.; Read, T.D.; Jerse, A.E.; Shafer, W.M. Phase-variable expression of lptA modulates the resistance of Neisseria gonorrhoeae to cationic antimicrobial peptides. Antimicrob. Agents Chemother. 2014, 58, 4230–4233. [Google Scholar] [CrossRef] [Green Version]
- Abu El-Asrar, A.M.; Struyf, S.; Descamps, F.J.; Al-Obeidan, S.A.; Proost, P.; Van Damme, J.; Opdenakker, G.; Geboes, K. Chemokines and gelatinases in the aqueous humor of patients with active uveitis. Am. J. Ophthalmol. 2004, 138, 401–411. [Google Scholar] [CrossRef]
- Lewis, L.A.; Shafer, W.M.; Dutta Ray, T.; Ram, S.; Rice, P.A. Phosphoethanolamine residues on the lipid A moiety of Neisseria gonorrhoeae lipooligosaccharide modulate binding of complement inhibitors and resistance to complement killing. Infect. Immun. 2013, 81, 33–42. [Google Scholar] [CrossRef] [Green Version]
- Handing, J.W.; Criss, A.K. The lipooligosaccharide-modifying enzyme LptA enhances gonococcal defence against human neutrophils. Cell. Microbiol. 2015, 17, 910–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Packiam, M.; Yedery, R.D.; Begum, A.A.; Carlson, R.W.; Ganguly, J.; Sempowski, G.D.; Ventevogel, M.S.; Shafer, W.M.; Jerse, A.E. Phosphoethanolamine decoration of Neisseria gonorrhoeae lipid A plays a dual immunostimulatory and protective role during experimental genital tract infection. Infect. Immun. 2014, 82, 2170–2179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hobbs, M.M.; Anderson, J.E.; Balthazar, J.T.; Kandler, J.L.; Carlson, R.W.; Ganguly, J.; Begum, A.A.; Duncan, J.A.; Lin, J.T.; Sparling, P.F.; et al. Lipid A’s structure mediates neisseria gonorrhoeae fitness during experimental infection of mice and men. mBio 2013, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergman, P.; Johansson, L.; Asp, V.; Plant, L.; Gudmundsson, G.H.; Jonsson, A.B.; Agerberth, B. Neisseria gonorrhoeae downregulates expression of the human antimicrobial peptide LL-37. Cell. Microbiol. 2005, 7, 1009–1017. [Google Scholar] [CrossRef]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef]
- Duan, Q.; Chen, H.; Costa, M.; Dai, W. Phosphorylation of H3S10 blocks the access of H3K9 by specific antibodies and histone methyltransferase. Implication in regulating chromatin dynamics and epigenetic inheritance during mitosis. J. Biol. Chem. 2008, 283, 33585–33590. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Duan, Y. Effects of posttranslational modifications on the structure and dynamics of histone H3 N-terminal peptide. Biophys. J. 2008, 94, 4579–4585. [Google Scholar] [CrossRef] [Green Version]
- Finnin, M.S.; Donigian, J.R.; Cohen, A.; Richon, V.M.; Rifkind, R.A.; Marks, P.A.; Breslow, R.; Pavletich, N.P. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 1999, 401, 188–193. [Google Scholar] [CrossRef]
- Moser, M.A.; Hagelkruys, A.; Seiser, C. Transcription and beyond: The role of mammalian class I lysine deacetylases. Chromosoma 2014, 123, 67–78. [Google Scholar] [CrossRef] [Green Version]
- Hildmann, C.; Ninkovic, M.; Dietrich, R.; Wegener, D.; Riester, D.; Zimmermann, T.; Birch, O.M.; Bernegger, C.; Loidl, P.; Schwienhorst, A. A new amidohydrolase from Bordetella or Alcaligenes strain FB188 with similarities to histone deacetylases. J. Bacteriol. 2004, 186, 2328–2339. [Google Scholar] [CrossRef] [Green Version]
- Iglesias, M.J.; Reilly, S.J.; Emanuelsson, O.; Sennblad, B.; Pirmoradian Najafabadi, M.; Folkersen, L.; Malarstig, A.; Lagergren, J.; Eriksson, P.; Hamsten, A.; et al. Combined chromatin and expression analysis reveals specific regulatory mechanisms within cytokine genes in the macrophage early immune response. PLoS ONE 2012, 7, e32306. [Google Scholar]
- Kumar, H.; Kawai, T.; Akira, S. Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 2009, 388, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 2009, 21, 317–337. [Google Scholar] [CrossRef] [Green Version]
- van der Meer, J.W.; Joosten, L.A.; Riksen, N.; Netea, M.G. Trained immunity: A smart way to enhance innate immune defence. Mol. Immunol. 2015, 68, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Blok, B.A.; Arts, R.J.; van Crevel, R.; Benn, C.S.; Netea, M.G. Trained innate immunity as underlying mechanism for the long-term, nonspecific effects of vaccines. J. Leukoc. Biol. 2015, 98, 347–356. [Google Scholar] [CrossRef]
- Ifrim, D.C.; Quintin, J.; Meerstein-Kessel, L.; Plantinga, T.S.; Joosten, L.A.; van der Meer, J.W.; van de Veerdonk, F.L.; Netea, M.G. Defective trained immunity in patients with STAT-1-dependent chronic mucocutaneaous candidiasis. Clin. Exp. Immunol. 2015, 181, 434–440. [Google Scholar] [CrossRef] [Green Version]
- Kleinnijenhuis, J.; van Crevel, R.; Netea, M.G. Trained immunity: Consequences for the heterologous effects of BCG vaccination. Trans. R. Soc. Trop. Med. Hyg. 2015, 109, 29–35. [Google Scholar] [CrossRef]
- Walker, C.K.; Sweet, R.L. Gonorrhea infection in women: Prevalence, effects, screening, and management. Int. J. Women’s Health 2011, 3, 197–206. [Google Scholar]
- Vodstrcil, L.A.; Fairley, C.K.; Fehler, G.; Leslie, D.; Walker, J.; Bradshaw, C.S.; Hocking, J.S. Trends in chlamydia and gonorrhea positivity among heterosexual men and men who have sex with men attending a large urban sexual health service in Australia, 2002–2009. BMC Infect. Dis. 2011, 11, 158. [Google Scholar] [CrossRef] [Green Version]
- Isabella, V.M.; Clark, V.L. Deep sequencing-based analysis of the anaerobic stimulon in Neisseria gonorrhoeae. BMC Genom. 2011, 12, 51. [Google Scholar] [CrossRef] [Green Version]
- Jerse, A.E.; Bash, M.C.; Russell, M.W. Vaccines against gonorrhea: Current status and future challenges. Vaccine 2014, 32, 1579–1587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolando, M.; Sanulli, S.; Rusniok, C.; Gomez-Valero, L.; Bertholet, C.; Sahr, T.; Margueron, R.; Buchrieser, C. Legionella pneumophila effector RomA uniquely modifies host chromatin to repress gene expression and promote intracellular bacterial replication. Cell Host Microbe 2013, 13, 395–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crotty Alexander, L.E.; Marsh, B.J.; Timmer, A.M.; Lin, A.E.; Zainabadi, K.; Czopik, A.; Guarente, L.; Nizet, V. Myeloid cell sirtuin-1 expression does not alter host immune responses to Gram-negative endotoxemia or Gram-positive bacterial infection. PLoS ONE 2013, 8, e84481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borutinskaite, V.; Navakauskiene, R. The histone deacetylase inhibitor BML-210 influences gene and protein expression in human promyelocytic leukemia NB4 cells via epigenetic reprogramming. Int. J. Mol. Sci. 2015, 16, 18252–18269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kallsen, K.; Andresen, E.; Heine, H. Histone deacetylase (HDAC) 1 controls the expression of beta defensin 1 in human lung epithelial cells. PLoS ONE 2012, 7, e50000. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Garcia, J.C.; Barat, N.C.; Trembley, S.J.; Dumler, J.S. Epigenetic silencing of host cell defense genes enhances intracellular survival of the rickettsial pathogen Anaplasma phagocytophilum. PLoS Pathog. 2009, 5, e1000488. [Google Scholar] [CrossRef]
- Garcia-Garcia, J.C.; Rennoll-Bankert, K.E.; Pelly, S.; Milstone, A.M.; Dumler, J.S. Silencing of host cell CYBB gene expression by the nuclear effector AnkA of the intracellular pathogen Anaplasma phagocytophilum. Infect. Immun. 2009, 77, 2385–2391. [Google Scholar] [CrossRef] [Green Version]
- Bierne, H.; Hamon, M.; Cossart, P. Epigenetics and bacterial infections. Cold Spring Harb. Perspect. Med. 2012, 2, a010272. [Google Scholar] [CrossRef]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Kelley, L.A.; Sternberg, M.J. Protein structure prediction on the Web: A case study using the phyre server. Nat. Protoc. 2009, 4, 363–371. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skaar, E.P.; Lecuyer, B.; Lenich, A.G.; Lazio, M.P.; Perkins-Balding, D.; Seifert, H.S.; Karls, A.C. Analysis of the piv recombinase-related gene family of Neisseria gonorrhoeae. J. Bacteriol. 2005, 187, 1276–1286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shafer, W.M.; Joiner, K.; Guymon, L.F.; Cohen, M.S.; Sparling, P.F. Serum sensitivity of Neisseria gonorrhoeae: The role of lipopolysaccharide. J. Infect. Dis. 1984, 149, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Jerse, A.E.; Sharma, N.D.; Simms, A.N.; Crow, E.T.; Snyder, L.A.; Shafer, W.M. A gonococcal efflux pump system enhances bacterial survival in a female mouse model of genital tract infection. Infect. Immun. 2003, 71, 5576–5582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouquette, C.; Harmon, J.B.; Shafer, W.M. Induction of the mtrCDE-encoded efflux pump system of Neisseria gonorrhoeae requires MtrA, an AraC-like protein. Mol. Microbiol. 1999, 33, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Crabb, D.M.; Dai, Y.; Chen, Y.; Waites, K.B.; Atkinson, T.P. Suppression of antimicrobial peptide expression by ureaplasma species. Infect. Immun. 2014, 82, 1657–1665. [Google Scholar] [CrossRef] [Green Version]
- Zughaier, S.M. Neisseria meningitidis capsular polysaccharides induce inflammatory responses via TLR2 and TLR4-MD-2. J. Leukoc. Biol. 2011, 89, 469–480. [Google Scholar] [CrossRef] [Green Version]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zughaier, S.M.; Rouquette-Loughlin, C.E.; Shafer, W.M. Identification of a Neisseria gonorrhoeae Histone Deacetylase: Epigenetic Impact on Host Gene Expression. Pathogens 2020, 9, 132. https://doi.org/10.3390/pathogens9020132
Zughaier SM, Rouquette-Loughlin CE, Shafer WM. Identification of a Neisseria gonorrhoeae Histone Deacetylase: Epigenetic Impact on Host Gene Expression. Pathogens. 2020; 9(2):132. https://doi.org/10.3390/pathogens9020132
Chicago/Turabian StyleZughaier, Susu M., Corinne E. Rouquette-Loughlin, and William M. Shafer. 2020. "Identification of a Neisseria gonorrhoeae Histone Deacetylase: Epigenetic Impact on Host Gene Expression" Pathogens 9, no. 2: 132. https://doi.org/10.3390/pathogens9020132