Coordination of Candida albicans Invasion and Infection Functions by Phosphoglycerol Phosphatase Rhr2
Abstract
:1. Introduction
2. Results
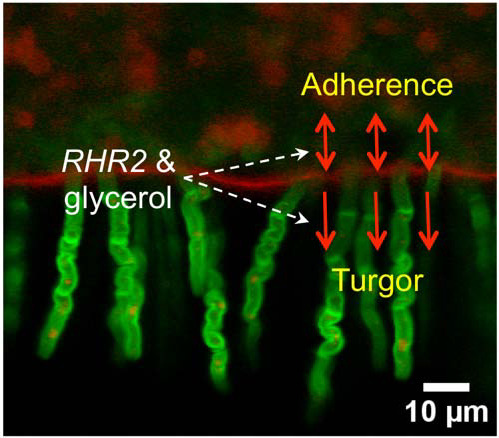
2.1. Mechanical and Genetic Determinants of Substrate Invasion

| Gene | Functional Description Form the Candida Genome Database | Mutant Phenotype |
|---|---|---|
| BRG1 | Transcription factor; recruits Hda1 to hypha-specific promoters; Tn mutation affects filamentation; Hap43-repressed; Spider and flow model biofilm induced; required for Spider biofilm formation; Bcr1-repressed in RPMI a/a biofilms | defective in biofilm formation and invasive growth into polyacrylamide |
| TEC1 | TEA/ATTS transcription factor; white cell pheromone response, hyphal gene regulation; required for Spider and RPMI biofilm formation; regulates BCR1; Cph2 regulated transcript; alkaline, rat catheter, Spider, flow model biofilm induced | |
| NDT80 | Ortholog of Ndt80; meiosis-specific transcription factor; activator of CDR1 induction by antifungal drugs; required for wild-type drug resistance and for Spider biofilm formation; transcript induced by antifungal drug treatment | |
| ROB1 | Zn(II)2Cys6 transcription factor; required for Spider model biofilm formation; mutant displays abnormal colony morphology and invasive growth; caspofungin repressed; flow model biofilm induced; rat catheter biofilm repressed | |
| DPB4 | Putative DNA polymerase epsilon subunit D; null mutant is viable but slow-growing and displays abnormal invasive growth on SD and YPD media; Spider biofilm repressed | |
| EFG1 | bHLH transcription factor; required for white-phase cell type, RPMI and Spider biofilm formation, hyphal growth, cell-wall gene regulation; roles in adhesion, virulence; Cph1 and Efg1 have role in host cytokine response; binds E-box |
2.2. Distinct RHR2 Requirements for Biofilm Formation and Invasion


2.3. RHR2 Function in Invasive Infection

3. Discussion
4. Experimental Section
4.1. Media and Strain Construction
| Strain | Genotype | Source |
|---|---|---|
| DAY185 (Wild-type) | ura3Δ::λiimm434 HIS1::his1::hisG ARG4::URA3::arg4::hisG | [45] |
| ura3Δ::λiimm434 his1::hisG arg4::hisG | ||
| JVD005 (rhr2Δ/Δ) | ura3Δ::λimm434 arg4::hisG his1::hisG::pHIS1 rhr2::ARG4 | [21] |
| ura3Δ::λimm434 arg4::hisG his1::hisG rhr2::URA3 | ||
| JVD006 (rhr2Δ/Δ + pRHR2) | ura3Δ::λimm434 arg4::hisG his1::hisG::pHIS1-RHR2 rhr2::ARG4 | [21] |
| ura3Δ::λimm434 arg4::hisG his1::hisG rhr2::URA3 | ||
| JVD039 (rhr2Δ/Δ + BCR1-OE) | ura3Δ::λimm434 arg4::hisG his1::hisG::pHIS1 rhr2::ARG4 | [21] |
| ALS1::pAgTEF1-NAT1-AgTEF1UTR-TDH3-BCR1 | ||
| ura3Δ::λimm434 arg4::hisG his1::hisG rhr2::URA3 BCR1 | ||
| JVD065 (rhr2Δ/Δ + BRG1-OE) | ura3Δ::λimm434 arg4::hisG his1::hisG::pHIS1 rhr2::ARG4 | This study |
| ALS1::pAgTEF1-NAT1-AgTEF1UTR-TDH3-BRG1 | ||
| ura3Δ::λimm434 arg4::hisG his1::hisG rhr2::URA3 BRG1 | ||
| JVD051 (rhr2Δ/Δ + UME6-OE) | ura3Δ::λimm434 arg4::hisG his1::hisG::pHIS1 rhr2::ARG4 | This study |
| ALS1::pAgTEF1-NAT1-AgTEF1UTR-TDH3-UME6 | ||
| ura3Δ::λimm434 arg4::hisG his1::hisG rhr2::URA3 UME6 | ||
| JVD018 (rhr2Δ/Δ + ALS1-OE) | ura3Δ::λimm434 arg4::hisG his1::hisG::pHIS1 rhr2::ARG4 | [21] |
| ALS1::pAgTEF1-NAT1-AgTEF1UTR-TDH3-ALS1 | ||
| ura3Δ::λimm434 arg4::hisG his1::hisG rhr2::URA3 ALS1 |
4.2. RNA Sample Preparation
4.3. Quantitative RT PCR
4.4. Polyacrylamide Gels for Invasion Assay
4.5. Elastic Modulus Measurement
4.6. Confocal Imaging of Invasion
4.7. Murine Model of Intra-Abdominal Candidiasis (IAC)
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Supplementary Information
References
- Lai, Y.; Rosenshine, I.; Leong, J.M.; Frankel, G. Intimate host attachment: Enteropathogenic and enterohaemorrhagic escherichia coli. Cell. Microbiol. 2013, 15, 1796–1808. [Google Scholar] [PubMed]
- Johnson, E.M.; Gaddy, J.A.; Cover, T.L. Alterations in helicobacter pylori triggered by contact with gastric epithelial cells. Front. Cell. Infect. Microbiol. 2012, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Mikula, K.M.; Kolodziejczyk, R.; Goldman, A. Yersinia infection tools-characterization of structure and function of adhesins. Front. Cell. Infect. Microbiol. 2013, 2, 169. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Filler, S.G. Candida albicans als3, a multifunctional adhesin and invasin. Eukaryot. Cell. 2011, 10, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.D.; Denning, D.W.; Gow, N.A.; Levitz, S.M.; Netea, M.G.; White, T.C. Hidden killers: Human fungal infections. Sci. Transl. Med. 2012, 4, 165rv113. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, R.S.; Robbins, N.; Cowen, L.E. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol. Mol. Biol. Rev. 2011, 75, 213–267. [Google Scholar] [CrossRef] [PubMed]
- Sudbery, P.E. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 2011, 9, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Finkel, J.S.; Mitchell, A.P. Genetic control of Candida albicans biofilm development. Nat. Rev. Microbiol. 2011, 9, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Filler, S.G. Interactions of Candida albicans with epithelial cells. Cell. Microbiol. 2010, 12, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Desai, J.V.; Mitchell, A.P.; Andes, D.R. Fungal biofilms, drug resistance, and recurrent infection. Cold Spring Harb. Perspect. Med. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Brand, A. Hyphal growth in human fungal pathogens and its role in virulence. Int. J. Microbiol. 2012, 2012, 517529. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.N.; Solis, N.V.; Phan, Q.T.; Bajwa, J.S.; Kashleva, H.; Thompson, A.; Liu, Y.; Dongari-Bagtzoglou, A.; Edgerton, M.; Filler, S.G. Host cell invasion and virulence mediated by Candida albicans ssa1. PLoS Pathog. 2010, 6, e1001181. [Google Scholar] [CrossRef] [PubMed]
- Phan, Q.T.; Myers, C.L.; Fu, Y.; Sheppard, D.C.; Yeaman, M.R.; Welch, W.H.; Ibrahim, A.S.; Edwards, J.E., Jr.; Filler, S.G. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Boil. 2007, 5, e64. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Phan, Q.T.; Boontheung, P.; Solis, N.V.; Loo, J.A.; Filler, S.G. Egfr and her2 receptor kinase signaling mediate epithelial cell invasion by Candida albicans during oropharyngeal infection. Proc. Natl. Acad. Sci. USA 2012, 109, 14194–14199. [Google Scholar] [CrossRef] [PubMed]
- Leonhard, M.; Moser, D.; Reumueller, A.; Mancusi, G.; Bigenzahn, W.; Schneider-Stickler, B. Comparison of biofilm formation on new phonax and provox 2 voice prostheses—A pilot study. Head Neck 2010, 32, 886–895. [Google Scholar] [CrossRef] [PubMed]
- Bastmeyer, M.; Deising, H.B.; Bechinger, C. Force exertion in fungal infection. Ann. Rev. Biophys. Biomol. Struct. 2002, 31, 321–341. [Google Scholar] [CrossRef] [PubMed]
- Almeida, R.S.; Wilson, D.; Hube, B. Candida albicans iron acquisition within the host. FEMS Yeast Res. 2009, 9, 1000–1012. [Google Scholar] [CrossRef] [PubMed]
- Bechinger, C.; Giebel, K.F.; Schnell, M.; Leiderer, P.; Deising, H.B.; Bastmeyer, M. Optical measurements of invasive forces exerted by appressoria of a plant pathogenic fungus. Science 1999, 285, 1896–1899. [Google Scholar] [CrossRef] [PubMed]
- Howard, R.J.; Ferrari, M.A.; Roach, D.H.; Money, N.P. Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl. Acad. Sci. USA 1991, 88, 11281–11284. [Google Scholar] [CrossRef] [PubMed]
- Bonhomme, J.; Chauvel, M.; Goyard, S.; Roux, P.; Rossignol, T.; d’Enfert, C. Contribution of the glycolytic flux and hypoxia adaptation to efficient biofilm formation by Candida albicans. Mol. Microbiol. 2011, 80, 995–1013. [Google Scholar] [CrossRef] [PubMed]
- Desai, J.V.; Bruno, V.M.; Ganguly, S.; Stamper, R.J.; Mitchell, K.F.; Solis, N.; Hill, E.M.; Xu, W.; Filler, S.G.; Andes, D.R.; et al. Regulatory role of glycerol in Candida albicans biofilm formation. mBio 2013, 4, e00637–00612. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wang, H.; Shang, Q.; Jiang, Y.; Cao, Y.; Chai, Y. Time course analysis of Candida albicans metabolites during biofilm development. J. Proteome Res. 2013, 12, 2375–2385. [Google Scholar] [CrossRef] [PubMed]
- Wachtler, B.; Wilson, D.; Haedicke, K.; Dalle, F.; Hube, B. From attachment to damage: Defined genes of Candida albicans mediate adhesion, invasion and damage during interaction with oral epithelial cells. PLoS ONE 2011, 6, e17046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, D.H., Jr.; Giusani, A.D.; Chen, X.; Kumamoto, C.A. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique czf1 gene. Mol. Microbiol. 1999, 34, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Riggle, P.J.; Andrutis, K.A.; Chen, X.; Tzipori, S.R.; Kumamoto, C.A. Invasive lesions containing filamentous forms produced by a Candida albicans mutant that is defective in filamentous growth in culture. Infect. Immun. 1999, 67, 3649–3652. [Google Scholar] [PubMed]
- Wachtler, B.; Citiulo, F.; Jablonowski, N.; Forster, S.; Dalle, F.; Schaller, M.; Wilson, D.; Hube, B. Candida albicans-epithelial interactions: Dissecting the roles of active penetration, induced endocytosis and host factors on the infection process. PLoS ONE 2012, 7, e36952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tucker, S.L.; Talbot, N.J. Surface attachment and pre-penetration stage development by plant pathogenic fungi. Ann. Rev. Phytopathol. 2001, 39, 385–417. [Google Scholar] [CrossRef] [PubMed]
- Homann, O.R.; Dea, J.; Noble, S.M.; Johnson, A.D. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009, 5, e1000783. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; VandeWalle, K.; Lopez-Ribot, J.L.; Wickes, B.L. The filamentation pathway controlled by the efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol. Lett. 2002, 214, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Mitchell, A.P. Regulation of cell-surface genes and biofilm formation by the c. Albicans transcription factor bcr1p. Curr. Boil.: CB 2005, 15, 1150–1155. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Fox, E.P.; Nett, J.E.; Sorrells, T.R.; Mitrovich, Q.M.; Hernday, A.D.; Tuch, B.B.; Andes, D.R.; Johnson, A.D. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 2012, 148, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Guan, G.; Xie, J.; Sun, Y.; Tong, Y.; Zhang, L.; Huang, G. Roles of Candida albicans gat2, a gata-type zinc finger transcription factor, in biofilm formation, filamentous growth and virulence. PLoS ONE 2012, 7, e29707. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Jenkinson, J.M.; Holcombe, L.J.; Soanes, D.M.; Veneault-Fourrey, C.; Bhambra, G.K.; Talbot, N.J. The molecular biology of appressorium turgor generation by the rice blast fungus magnaporthe grisea. Biochem. Soc. Trans. 2005, 33, 384–388. [Google Scholar] [PubMed]
- Cheng, S.; Clancy, C.J.; Xu, W.; Schneider, F.; Hao, B.; Mitchell, A.P.; Nguyen, M.H. Profiling of Candida albicans gene expression during intra-abdominal candidiasis identifies biologic processes involved in pathogenesis. J. Infect. Dis. 2013, 208, 1529–1537. [Google Scholar] [CrossRef] [PubMed]
- Ell, S.R. Candida—‘The Cancer of Silastic’. J. Laryngol. Otol. 1996, 110, 240–242. [Google Scholar] [CrossRef] [PubMed]
- Van Weissenbruch, R.; Albers, F.W.; Bouckaert, S.; Nelis, H.J.; Criel, G.; Remon, J.P.; Sulter, A.M. Deterioration of the provox silicone tracheoesophageal voice prosthesis: Microbial aspects and structural changes. Acta Otolaryngol. 1997, 117, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.D.; Wehmeier, S.; Byfield, F.J.; Janmey, P.A.; Caballero-Lima, D.; Crossley, A.; Brand, A.C. Contact-induced apical asymmetry drives the thigmotropic responses of Candida albicans hyphae. Cell. Microbiol. 2015, 17, 342–354. [Google Scholar] [CrossRef] [PubMed]
- Fanning, S.; Xu, W.; Solis, N.; Woolford, C.A.; Filler, S.G.; Mitchell, A.P. Divergent targets of Candida albicans biofilm regulator bcr1 in vitro and in vivo. Eukaryot. Cell 2012, 11, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Yeh, W.C.; Li, P.C.; Jeng, Y.M.; Hsu, H.C.; Kuo, P.L.; Li, M.L.; Yang, P.M.; Lee, P.H. Elastic modulus measurements of human liver and correlation with pathology. Ultrasound Med. Boil. 2002, 28, 467–474. [Google Scholar] [CrossRef]
- Wiese, K.G. Electrolyte concentration, real and osmotic pressure in abscesses. Zentralblatt Fur Chir. 1994, 119, 54–59. [Google Scholar]
- Fan, J.; Whiteway, M.; Shen, S.H. Disruption of a gene encoding glycerol 3-phosphatase from Candida albicans impairs intracellular glycerol accumulation-mediated salt-tolerance. FEMS Microbiol. Lett. 2005, 245, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Boil. Rev.: MMBR 2002, 66, 300–372. [Google Scholar] [CrossRef]
- Pahlman, A.K.; Granath, K.; Ansell, R.; Hohmann, S.; Adler, L. The yeast glycerol 3-phosphatases gpp1p and gpp2p are required for glycerol biosynthesis and differentially involved in the cellular responses to osmotic, anaerobic, and oxidative stress. J. Boil. Chem. 2001, 276, 3555–3563. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Fabrizio, P.; Madia, F.; Hu, J.; Ge, H.; Li, L.M.; Longo, V.D. Tor1/sch9-regulated carbon source substitution is as effective as calorie restriction in life span extension. PLoS Genet. 2009, 5, e1000467. [Google Scholar] [CrossRef] [PubMed]
- Davis, D.; Edwards, J.E., Jr.; Mitchell, A.P.; Ibrahim, A.S. Candida albicans rim101 ph response pathway is required for host-pathogen interactions. Infect. Immun. 2000, 68, 5953–5959. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Desai, J.V.; Cheng, S.; Ying, T.; Nguyen, M.H.; Clancy, C.J.; Lanni, F.; Mitchell, A.P. Coordination of Candida albicans Invasion and Infection Functions by Phosphoglycerol Phosphatase Rhr2. Pathogens 2015, 4, 573-589. https://doi.org/10.3390/pathogens4030573
Desai JV, Cheng S, Ying T, Nguyen MH, Clancy CJ, Lanni F, Mitchell AP. Coordination of Candida albicans Invasion and Infection Functions by Phosphoglycerol Phosphatase Rhr2. Pathogens. 2015; 4(3):573-589. https://doi.org/10.3390/pathogens4030573
Chicago/Turabian StyleDesai, Jigar V., Shaoji Cheng, Tammy Ying, M. Hong Nguyen, Cornelius J. Clancy, Frederick Lanni, and Aaron P. Mitchell. 2015. "Coordination of Candida albicans Invasion and Infection Functions by Phosphoglycerol Phosphatase Rhr2" Pathogens 4, no. 3: 573-589. https://doi.org/10.3390/pathogens4030573