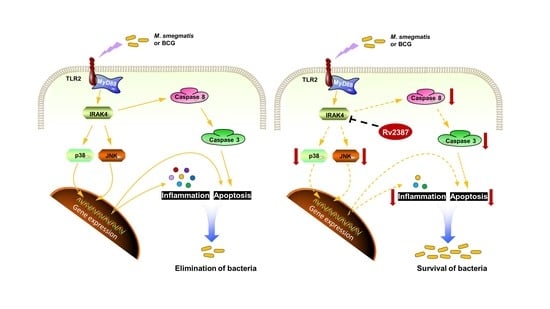
Mycobacterium tuberculosis Rv2387 Facilitates Mycobacterial Survival by Silencing TLR2/p38/JNK Signaling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria, Cell Lines, and Animals
2.2. Construction of Stable Cell Line
2.3. Isolation of Mouse Murine Bone Marrow-Derived Macrophages (BMDMs)
2.4. Intracellular Survival Assay
2.5. Real-Time Quantitative PCR Assay
2.6. ELISA Assay
2.7. Lactate Dehydrogenase (LDH) Determination
2.8. Necrosis Assay
2.9. Flow Cytometry
2.10. TUNEL Assay
2.11. Western Blot
2.12. Evaluation of the Virulence of Msmg-2387 in Mice
2.13. Statistical Analysis
3. Results
3.1. Rv2387 Enhanced the Intracellular Survival of Recombinant Msmg-2387 in Macrophages
3.2. Rv2387 Suppressed Host Inflammatory Response to Mycobacterial Infection
3.3. Rv2387 Blocked M. Smegmatis-Induced Macrophage Apoptosis
3.4. Rv2387 Inhibited Apoptosis by Suppressing the Extrinsic Apoptotic Pathway
3.5. Rv2387 Inhibited M. Smegmatis-Induced p38 and JNK Activation
3.6. The Effect of Rv2387 on Suppressing p38 and JNK Activation was Dependent on TLR2 Signaling
3.7. The Role of Rv2387 Protein Was Evaluated in BMDM Cells and Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Gobal Tuberculosis Report 2021. Available online: https://www.who.int/publications/i/item/9789240037021 (accessed on 14 October 2021).
- Pai, M.; Kasaeva, T.; Swaminathan, S. COVID-19’s Devastating Effect on Tuberculosis Care—A Path to Recovery. N. Engl. J. Med. 2022, 386, 1490–1493. [Google Scholar] [CrossRef] [PubMed]
- Ly, A.; Liu, J. Mycobacterial Virulence Factors: Surface-Exposed Lipids and Secreted Proteins. Int. J. Mol. Sci. 2020, 21, 3985. [Google Scholar] [CrossRef] [PubMed]
- de Martino, M.; Lodi, L.; Galli, L.; Chiappini, E. Immune Response to Mycobacterium tuberculosis: A Narrative Review. Front. Pediatr. 2019, 7, 350. [Google Scholar] [CrossRef] [PubMed]
- Briken, V.; Miller, J.L. Living on the edge: Inhibition of host cell apoptosis by Mycobacterium tuberculosis. Future Microbiol. 2008, 3, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.; Mittal, E.; Philips, J.A. Tuberculosis and the art of macrophage manipulation. Pathog. Dis. 2018, 76. [Google Scholar] [CrossRef] [PubMed]
- Bohsali, A.; Abdalla, H.; Velmurugan, K.; Briken, V. The non-pathogenic mycobacteria M. smegmatis and M. fortuitum induce rapid host cell apoptosis via a caspase-3 and TNF dependent pathway. BMC Microbiol. 2010, 10, 237. [Google Scholar] [CrossRef]
- Feng, Z.H.; Bai, X.Y.; Wang, T.; Garcia, C.; Bai, A.; Li, L.; Honda, J.R.; Nie, X.H.; Chan, E.W.D. Differential responses by human macrophages to infection with Mycobacterium tuberculosis and non-tuberculous mycobacteria. Front. Microbiol. 2020, 11, 116. [Google Scholar] [CrossRef]
- Lam, A.; Prabhu, R.; Gross, C.M.; Riesenberg, L.A.; Singh, V.; Aggarwal, S. Role of apoptosis and autophagy in tuberculosis. Am. J. Physiol. Cell. Mol. Physiol. 2017, 313, L218–L229. [Google Scholar] [CrossRef]
- Liu, C.H.; Liu, H.; Ge, B. Innate immunity in tuberculosis: Host defense vs. pathogen evasion. Cell Mol. Immunol. 2017, 14, 963–975. [Google Scholar] [CrossRef]
- Zhang, W.; Lu, Q.; Dong, Y.; Yue, Y.; Xiong, S. Rv3033, as an Emerging Anti-apoptosis Factor, Facilitates Mycobacteria Survival via Inhibiting Macrophage Intrinsic Apoptosis. Front. Immunol. 2018, 9, 2136. [Google Scholar] [CrossRef] [Green Version]
- Edwards, K.M.; Cynamon, M.H.; Voladri, R.K.R.; Hager, C.C.; DeStefano, M.S.; Tham, K.T.; Lakey, D.L.; Bochan, M.R.; Kernodle, D.S. Iron-cofactored superoxide dismutase inhibits host responses to Mycobacterium tuberculosis. Am. J. Respir. Crit. Care Med. 2001, 164, 2213–2219. [Google Scholar] [CrossRef] [PubMed]
- Velmurugan, K.; Chen, B.; Miller, J.L.; Azogue, S.; Gurses, S.; Hsu, T.; Glickman, M.; Jacobs, W.R.; Porcelli, S.A.; Briken, V. Mycobacterium tuberculosis nuoG is a virulence gene that Inhibits apoptosis of infected host cells. PLoS Pathog. 2007, 3, e110. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.L.; Velmurugan, K.; Cowan, M.J.; Briken, V. The type I NADH dehydrogenase of Mycobacterium tuberculosis counters phagosomal NOX2 activity to inhibit TNF-alpha-mediated host cell apoptosis. PLoS Pathog. 2010, 6, e1000864. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.M.; Jeon, B.Y.; Lee, H.M.; Jin, H.S.; Yuk, J.M.; Song, C.H.; Lee, S.H.; Lee, Z.W.; Cho, S.N.; Kim, J.M.; et al. Mycobacterium tuberculosis eis regulates autophagy, inflammation, and cell death through redox-dependent signaling. PLoS Pathog. 2010, 6, e1001230. [Google Scholar] [CrossRef] [PubMed]
- Danelishvili, L.; Everman, J.L.; McNamara, M.J.; Bermudez, L.E. Inhibition of the Plasma-Membrane-Associated Serine Protease Cathepsin G by Mycobacterium tuberculosis Rv3364c Suppresses Caspase-1 and Pyroptosis in Macrophages. Front. Microbiol. 2012, 2. [Google Scholar] [CrossRef]
- Kapopoulou, A.; Lew, J.M.; Cole, S.T. The MycoBrowser portal: A comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis 2011, 91, 8–13. [Google Scholar] [CrossRef]
- Li, W.; Fan, X.; Long, Q.; Xie, L.; Xie, J. Mycobacterium tuberculosis effectors involved in host-pathogen interaction revealed by a multiple scales integrative pipeline. Infect. Genet. Evol. 2015, 32, 1–11. [Google Scholar] [CrossRef]
- Alibaud, L.; Rombouts, Y.; Trivelli, X.; Burguière, A.; Cirillo, S.L.G.; Cirillo, J.D.; Dubremetz, J.-F.; Guérardel, Y.; Lutfalla, G.; Kremer, L. A Mycobacterium marinum TesA mutant defective for major cell wall-associated lipids is highly attenuated in Dictyostelium discoideum and zebrafish embryos. Mol. Microbiol. 2011, 80, 919–934. [Google Scholar] [CrossRef]
- Jin, C.; Wu, X.; Dong, C.; Li, F.; Fan, L.; Xiong, S.; Dong, Y. EspR promotes mycobacteria survival in macrophages by inhibiting MyD88 mediated inflammation and apoptosis. Tuberculosis 2019, 116, 22–31. [Google Scholar] [CrossRef]
- Qu, Z.; Zhou, J.; Zhou, Y.; Xie, Y.; Jiang, Y.; Wu, J.; Luo, Z.; Liu, G.; Yin, L.; Zhang, X.L. Mycobacterial EST12 activates a RACK1-NLRP3-gasdermin D pyroptosis-IL-1beta immune pathway. Sci Adv. 2020, 6, eaba4733. [Google Scholar] [CrossRef] [PubMed]
- Dheenadhayalan, V.; Delogu, G.; Brennan, M.J. Expression of the PE_PGRS 33 protein in Mycobacterium smegmatis triggers necrosis in macrophages and enhanced mycobacterial survival. Microbes Infect. 2006, 8, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.K.; Naqvi, N.; Alam, A.; Ahmad, J.; Alsati, B.S.; Sheikh, J.A.; Kumar, P.; Mitra, D.K.; Rahman, S.A.; Hasnain, S.E.; et al. Mycobacterium smegmatis bacteria expressing mycobacterium tuberculosis-specific Rv1954A induce macrophage activation and modulate the immune response. Front. Cell. Infect. Microbiol. 2020, 10, 564565. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Ma, C.Y.; Yan, Z.F.; Deng, R.; Ai, X.F.; Su, T.; Xiang, X.H.; Li, W. The Mycobacterium tuberculosis protein Rv2387 is involved in cell wall remodeling and susceptibility to acidic conditions. Biochem. Biophys. Res. Commun. 2018, 503, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Bashiri, G.; Perkowski, E.F.; Turner, A.P.; Feltcher, M.E.; Braunstein, M.; Baker, E.N. Tat–dependent translocation of an F420–Binding Protein of Mycobacterium tuberculosis. PLoS ONE 2012, 7, e45003. [Google Scholar] [CrossRef] [PubMed]
- Awuh, J.A.; Flo, T.H. Molecular basis of mycobacterial survival in macrophages. Cell Mol. Life Sci. 2017, 74, 1625–1648. [Google Scholar] [CrossRef]
- Li, W.; Zhao, Q.; Deng, W.; Chen, T.; Liu, M.; Xie, J. Mycobacterium tuberculosis Rv3402c Enhances Mycobacterial Survival within Macrophages and Modulates the Host Pro-Inflammatory Cytokines Production via NF-Kappa B/ERK/p38 Signaling. PLoS ONE 2014, 9, e94418. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.; Wang, D.; Li, M.; Wang, H.; Yu, J.; Wang, C.; Liu, J.; Gao, Q. PPE38 modulates the innate immune response and is required for Mycobacterium marinum virulence. Infect. Immun. 2012, 80, 43–54. [Google Scholar] [CrossRef]
- Behar, S.M.; Martin, C.J.; Booty, M.G.; Nishimura, T.; Zhao, X.; Gan, H.; Divangahi, M.; Remold, H.G. Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal Immunol. 2011, 4, 279–287. [Google Scholar] [CrossRef]
- Liu, M.; Li, W.; Xiang, X.; Xie, J. Mycobacterium tuberculosis effectors interfering host apoptosis signaling. Apoptosis 2015, 20, 883–891. [Google Scholar] [CrossRef]
- Lu, Q.; Zhang, W.; Fang, J.; Zheng, J.; Dong, C.; Xiong, S. Mycobacterium tuberculosis Rv1096, facilitates mycobacterial survival by modulating the NF-kappaB/MAPK pathway as peptidoglycan N-deacetylase. Mol. Immunol. 2020, 127, 47–55. [Google Scholar] [CrossRef]
- Li, F.; Feng, L.; Jin, C.; Wu, X.; Fan, L.; Xiong, S.; Dong, Y. LpqT improves mycobacteria survival in macrophages by inhibiting TLR2 mediated inflammatory cytokine expression and cell apoptosis. Tuberculosis 2018, 111, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Yang, C.S.; Shin, A.R.; Jeon, S.R.; Park, J.K.; Kim, H.J.; Jo, E.K. Mycobacterial Heparin-binding Hemagglutinin Antigen Activates Inflammatory Responses through PI3-K/Akt, NF-kappaB, and MAPK Pathways. Immune Netw. 2011, 11, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Paik, S.; Choi, S.; Lee, K.I.; Back, Y.W.; Son, Y.J.; Jo, E.K.; Kim, H.J. Mycobacterium tuberculosis acyl carrier protein inhibits macrophage apoptotic death by modulating the reactive oxygen species/c-Jun N-terminal kinase pathway. Microbes Infect. 2019, 21, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhu, X.; Peng, Y.; Zhu, T.; Liu, H.; Zhu, Y.; Xiong, X.; Chen, X.; Hu, C.; Chen, H.; et al. Mycobacterium tuberculosis YrbE3A Promotes Host Innate Immune Response by Targeting NF-kappaB/JNK Signaling. Microorganisms 2020, 8, 584. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xie, J. Role of mycobacteria effectors in phagosome maturation blockage and new drug targets discovery. J. Cell Biochem. 2011, 112, 2688–2693. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Silwal, P.; Jo, E.-K. Host-Pathogen Dialogues in Autophagy, Apoptosis, and Necrosis during Mycobacterial Infection. Immune Network. 2020, 20, e37. [Google Scholar] [CrossRef]
- Teo, W.; Yu, J.; Gao, Q. A novel method of screening for virulence genes from colony phenotype in Mycobacterium mariunm. Fudan Univ. J. Med. Sci. 2009, 36, 596–600. [Google Scholar]
- Danilchanka, O.; Sun, J.; Pavlenok, M.; Maueroder, C.; Speer, A.; Siroy, A.; Marrero, J.; Trujillo, C.; Mayhew, D.L.; Doornbos, K.S.; et al. An outer membrane channel protein of Mycobacterium tuberculosis with exotoxin activity. Proc. Natl. Acad. Sciences. 2014, 111, 6750–6755. [Google Scholar] [CrossRef]
- Sun, J.; Siroy, A.; Lokareddy, R.K.; Speer, A.; Doornbos, K.S.; Cingolani, G.; Niederweis, M. The tuberculosis necrotizing toxin kills macrophages by hydrolyzing NAD. Nat. Struct. Mol. Biol. 2015, 22, 672–678. [Google Scholar] [CrossRef]
- Danilchanka, O.; Pires, D.; Anes, E.; Niederweis, M. The Mycobacterium tuberculosis Outer Membrane Channel Protein CpnT Confers Susceptibility to Toxic Molecules. Antimicrob. Agents Chemother. 2015, 59, 2328–2336. [Google Scholar] [CrossRef]
- O’Leary, S.; O’Sullivan, M.P.; Keane, J. IL-10 Blocks Phagosome Maturation in Mycobacterium tuberculosis-infected Human Macrophages. Am. J. Respir. Cell Mol. Biol. 2011, 45, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Redford, P.S.; Boonstra, A.; Read, S.; Pitt, J.; Graham, C.; Stavropoulos, E.; Bancroft, G.J.; O’Garra, A. Enhanced protection to Mycobacterium tuberculosis infection in IL-10-deficient mice is accompanied by early and enhanced Th1 responses in the lung. Eur. J. Immunol. 2010, 40, 2200–2210. [Google Scholar] [CrossRef] [PubMed]
- Hwanga, E.H.; Kim, T.H.; Park, J.Y.; Hong, J.J.; Kim, D.H.; Ha, S.J.; Yang, S.J.; Shin, S.J.; Park, J.H. TLR2 contributes to trigger immune response of pleural mesothelial cells against Mycobacterium bovis BCG and M. tuberculosis infection. Cytokine 2017, 95, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Fremond, C.M.; Togbe, D.; Doz, E.; Rose, S.; Vasseur, V.; Maillet, I.; Jacobs, M.; Ryffel, B.; Quesniaux, V.F. IL-1 receptor-mediated signal is an essential component of MyD88-dependent innate response to Mycobacterium tuberculosis infection. J. Immunol. 2007, 179, 1178–1189. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, B.X.; Ge, P.P.; Li, J.; Wang, Q.; Gao, G.F.; Qiu, X.B.; Liu, C.H. Mycobacterium tuberculosis suppresses innate immunity by coopting the host ubiquitin system. Nat. Immunol. 2015, 16, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, D.; Rojas, M.; Hernandez, I.; Radzioch, D.; Garcia, L.F.; Barrera, L.F. Role of TLR2-and TLR4-mediated signaling in Mycobacterium tuberculosis-induced macrophage death. Cell. Immunol. 2010, 260, 128–136. [Google Scholar] [CrossRef]
- Romero, M.M.; Basile, J.I.; Feo, L.C.; Lopez, B.; Ritacco, V.; Aleman, M. Reactive oxygen species production by human dendritic cells involves TLR2 and dectin-1 and is essential for efficient immune response against Mycobacteria. Cell. Microbiol. 2016, 18, 875–886. [Google Scholar] [CrossRef]
- Zhai, W.; Wu, F.; Zhang, Y.; Fu, Y.; Liu, Z. The Immune Escape Mechanisms of Mycobacterium Tuberculosis. Int. J. Mol. Sci. 2019, 20, 340. [Google Scholar] [CrossRef]
- Behar, S.M.; Briken, V. Apoptosis inhibition by intracellular bacteria and its consequence on host immunity. Curr. Opin. Immunol. 2019, 60, 103–110. [Google Scholar] [CrossRef]
- Molloy, A.; Laochumroonvorapong, P.; Kaplan, G. Apoptosis, but not necrosis, of infected monocytes is coupled with killing of intracellular bacillus Calmette-Guerin. J. Exp. Med. 1994, 180, 1499–1509. [Google Scholar] [CrossRef]
- Keane, J.; Shurtleff, B.; Kornfeld, H. TNF-dependent BALB/c murine macrophage apoptosis following Mycobacterium tuberculosis infection inhibits bacillary growth in an IFN-gamma independent manner. Tuberculosis 2002, 82, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Keane, J.; Remold, H.G.; Kornfeld, H. Virulent Mycobacterium tuberculosis strains evade apoptosis of infected alveolar macrophages. J. Immunol. 2000, 164, 2016–2020. [Google Scholar] [CrossRef] [PubMed]
- Poirier, V.; Bach, H.; Av-Gay, Y. Mycobacterium tuberculosis Promotes Anti-apoptotic Activity of the Macrophage by PtpA Protein-dependent Dephosphorylation of Host GSK3alpha. J. Biol. Chem. 2014, 289, 29376–29385. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.; Yuen, A.; Singh, V.; Hmama, Z. Mycobacterium tuberculosis Cpn60.2 (GroEL2) blocks macrophage apoptosis via interaction with mitochondrial mortalin. Biol. Open 2017, 6, 481–488. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, S.; Tang, Y.; Liu, Q.; Yao, Y. MPT64 protein from Mycobacterium tuberculosis inhibits apoptosis of macrophages through NF-kB-miRNA21-Bcl-2 pathway. PLoS ONE 2014, 9, e100949. [Google Scholar] [CrossRef] [PubMed]
- Danelishvili, L.; Yamazaki, Y.; Selker, J.; Bermudez, L.E. Secreted Mycobacterium tuberculosis Rv3654c and Rv3655c proteins participate in the suppression of macrophage apoptosis. PLoS ONE 2010, 5, e10474. [Google Scholar] [CrossRef]







| Primers | Sequence |
|---|---|
| IL-10-Forw | ATGCCTGGCTCAGAC |
| IL-10-Rev | GTCCTGCATTAAGGAGTCG |
| IL-1β-Forw | GCAACTGTTCCTGAACTCAACT |
| IL-1β-Rev | ATCTTTTGGGGTCCGTCAACT |
| TNF-α-Forw | CCCTCACACTCAGATCATCTTCT |
| TNF-α-Rev | GCTACGACGTGGGCTACAG |
| IL-6-Forw | GAGAGGAGACTTCACAGAGGATAC |
| IL-6-Rev | GTACTCCAGAAGACCAGAGG |
| Gapdh-Forw | GAAGGGCTCATGACCACAGT |
| Gapdh-Rev | GGATGCAGGGATGATGTTCT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Deng, W.; Zhang, N.; Peng, H.; Xu, Y. Mycobacterium tuberculosis Rv2387 Facilitates Mycobacterial Survival by Silencing TLR2/p38/JNK Signaling. Pathogens 2022, 11, 981. https://doi.org/10.3390/pathogens11090981
Li W, Deng W, Zhang N, Peng H, Xu Y. Mycobacterium tuberculosis Rv2387 Facilitates Mycobacterial Survival by Silencing TLR2/p38/JNK Signaling. Pathogens. 2022; 11(9):981. https://doi.org/10.3390/pathogens11090981
Chicago/Turabian StyleLi, Wu, Wanyan Deng, Nan Zhang, Huijuan Peng, and Yi Xu. 2022. "Mycobacterium tuberculosis Rv2387 Facilitates Mycobacterial Survival by Silencing TLR2/p38/JNK Signaling" Pathogens 11, no. 9: 981. https://doi.org/10.3390/pathogens11090981