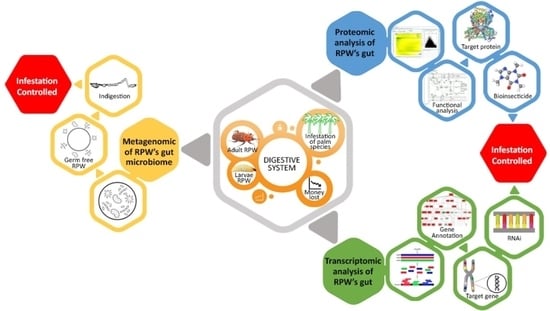
A Review on Digestive System of Rhynchophorus ferrugineus as Potential Target to Develop Control Strategies
Abstract
:Simple Summary
Abstract
1. Introduction
2. Anatomy of RPW’s Digestive System
3. Gut Bacteria of RPW
4. Protein Profile of RPW’s Gut
5. RPW’s Gut Transcriptome Analysis
6. R. ferrugineus Control Related to Its Digestive System
7. Opportunity and Challenges of Targeting RPW’s Digestive System
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hazzouri, K.M.; Sudalaimuthuasari, N.; Kundu, B.; Nelson, D.; Al-Deeb, M.A.; Le Mansour, A.; Spencer, J.J.; Desplan, C.; Amiri, K.M.A. The Genome of Pest Rhynchophorus ferrugineus Reveals Gene Families Important at the Plant-Beetle Interface. Commun. Biol. 2020, 3, 323. [Google Scholar] [CrossRef]
- Azmi, W.A.; Lian, C.J.; Zakeri, H.A.; Yusuf, N.; Omar, W.B.W.; Wai, Y.K.; Zulkefli, A.N.; Hussain, M.H. The Red Palm Weevil, Rhynchophorus ferrugineus: Current Issues and Challenges in Malaysia. Oil Palm. Bull. 2017, 74, 17–24. [Google Scholar]
- Harith-Fadzilah, N.; Haris-Hussain, M.; Abd Ghani, I.; Zakaria, A.; Amit, S.; Zainal, Z.; Azmi, W.A.; Jalinas, J.; Hassan, M. Physical and Physiological Monitoring on Red Palm Weevil-Infested Oil Palms. Insects 2020, 11, 407. [Google Scholar] [CrossRef]
- Ahmad, J.N.; Manzoor, M.; Aslam, Z.; Ahmad, S.J.N. Molecular Study on Field Evolved Resistance of Red Palm Weevil (Rhynchophorus Ferruginous) and Its Management through RNAi. PJZ 2020, 52, 477. [Google Scholar] [CrossRef]
- Siriwardena, K.A.P.; Fernando, L.C.P.; Nanayakkara, N.; Perera, K.F.G.; Kumara, A.D.N.T.; Nanayakkara, T. Portable Acoustic Device for Detection of Coconut Palms Infested by Rynchophorus ferrugineus (Coleoptera: Curculionidae). Crop. Prot. 2010, 29, 25–29. [Google Scholar] [CrossRef]
- Hussain, M.H. Efficacy of Baits for Red Palm Weevil (RPW), Rhynchophorus ferrugineus Olivier under Constant Laboratory Condition. JOPR 2020, 32, 355–364. [Google Scholar] [CrossRef]
- Soroker, V.; Colazza, S. Handbook of Major Palm Pests: Biology and Management; John Wiley & Sons: Hoboken, NJ, USA, 2017; ISBN 978-1-119-05745-1. [Google Scholar]
- Leatemia, J.A.; Patty, J.A.; Masauna, E.D.; Noya, S.H.; Hasinu, J.V. Utilization of Sago Grub (Rhynchophorus ferrugineus Olivier) (Coleoptera: Curculionidae) as an Alternative Source of Protein. IOP Conf. Ser. Earth Environ. Sci. 2021, 800, 012028. [Google Scholar] [CrossRef]
- El-Mergawy, R.; Al-Ajlan, A.M. Red Palm Weevil, Rhynchophorus ferrugineus (Olivier): Economic Importance, Biology, Biogeography and Integrated Pest Management. J. Agric. Sci. Technol. A 2011, 1, 1–23. [Google Scholar]
- Nurashikin-Khairuddin, W.; Abdul-Hamid, S.N.A.; Mansor, M.S.; Bharudin, I.; Othman, Z.; Jalinas, J. A Review of Entomopathogenic Nematodes as a Biological Control Agent for Red Palm Weevil, Rhynchophorus ferrugineus (Coleoptera: Curculionidae). Insects 2022, 13, 245. [Google Scholar] [CrossRef]
- Yang, H.; Xu, D.; Zhuo, Z.; Hu, J.; Lu, B. Transcriptome and Gene Expression Analysis of Rhynchophorus ferrugineus (Coleoptera: Curculionidae) during Developmental Stages. PeerJ 2020, 8, e10223. [Google Scholar] [CrossRef]
- Harris, M.N.; Norzainih, J.J.; Nurul Wahida, O. Morphology and Histology of the Digestive System of the Red Palm Weevil Larva, Rhynchophorus ferrugineus, Olivier (Coleoptera: Dryophthoridae). In Proceedings of the 3rd International Conference on Chemical, Agricultural and Medical Sciences (CAMS-2015), Singapore, 10–11 December 2015; International Institute of Chemical, Biological & Environmental Engineering: Phuket, Thailand, 2015; pp. 32–37. [Google Scholar]
- El-Fattah, A.Y.A.; El-Wahab, A.S.A.; Jamal, Z.A.; El-Helaly, A.A. Histopathological Studies of Red Palm Weevil Rhynchophorus ferrugineus, (Olivier) Larvae and Adults to Evaluate Certain Nano Pesticides. Braz. J. Biol. 2021, 81, 195–201. [Google Scholar] [CrossRef]
- El-Sobki, A.; Ali, A. Biochemical Effects of Some Chitin Synthesis Inhibitors Against Red Palm Weevil, Rhynchophorus ferrugineus Insect. Egypt. Acad. J. Biol. Sci. F. Toxicol. Pest Control 2020, 12, 127–139. [Google Scholar] [CrossRef]
- Harris, M.N.; Shafinaz, N.; Yaakop, S.; Othman, N.W. Distribution of Serotonin (5-HT) and Dopamine (DA) on Digestive Tract of Red Palm Weevil Larva, Rhynchophorus ferrugineus (Coleoptera: Dryophthoridae). Sains Malays 2016, 21, 39–50. [Google Scholar]
- Mohamed, M.A.; Shaalan, S.; Ghazy, A.-E.M.; Ali, A.A.; Abd-Elaziz, A.M.; Ghanem, M.M.E.; Abd-Elghany, S.A. Purification and Characterization of Acetylcholinesterase in Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae). Int. J. Biol. Macromol. 2020, 147, 1029–1040. [Google Scholar] [CrossRef]
- Wakil, W.; Yasin, M.; Qayyum, M.A.; Ghazanfar, M.U.; Al-Sadi, A.M.; Bedford, G.O.; Kwon, Y.J. Resistance to Commonly Used Insecticides and Phosphine Fumigant in Red Palm Weevil, Rhynchophorus ferrugineus (Olivier) in Pakistan. PLoS ONE 2018, 13, e0192628. [Google Scholar] [CrossRef]
- Hussain, A.; AlJabr, A.M.; Al-Ayedh, H. Development-Disrupting Chitin Synthesis Inhibitor, Novaluron, Reprogramming the Chitin Degradation Mechanism of Red Palm Weevils. Molecules 2019, 24, 4304. [Google Scholar] [CrossRef]
- Mahadi, N.A.; Yusof, T.A.A.; Mat, M.; Abdullah, A.; Zakaria, M.H.; Masdor, N.A. Comparative Study of Red Palm Weevil (RPW), Rhynchophorus ferrugineus (Coleoptera: Dryophthoridae) Reproduction Reared Using Different Diets. Am. J. Entomol. 2022, 6, 110–115. [Google Scholar] [CrossRef]
- Zulkifli, A.N.; Zakeri, H.A.; Azmi, W.A. Food Consumption, Developmental Time, and Protein Profile of the Digestive System of the Red Palm Weevil, Rhynchophorus ferrugineus (Coleptera: Dryophthoridae) Larvae Reared on Three Different Diets. J. Insect Sci. 2018, 18, 10. [Google Scholar] [CrossRef]
- Haldhar, S.M.; Muralidharan, C.M.; Singh, D. Pests and Their Management in Date Palm. In Trends in Horticultural Entomology; Springer: Singapore, 2022; pp. 833–845. [Google Scholar]
- Dembilio, Ó.; Jaques, J.A. Biology and Management of Red Palm Weevil. In Sustainable Pest Management in Date Palm: Current Status and Emerging Challenges; Wakil, W., Romeno Faleiro, J., Miller, T.A., Eds.; Sustainability in Plant and Crop Protection; Springer International Publishing: Cham, Switzerland, 2015; pp. 13–36. ISBN 978-3-319-24397-9. [Google Scholar]
- Martin, B. A Relative Analysis on Sound of Red Palm Weevil Based on Field and Lab Recordings. Int. J. Appl. Eng. Res. 2015, 10, 5261–5268. [Google Scholar]
- Muhammad, A.; Habineza, P.; Hou, Y.; Shi, Z. Preparation of Red Palm Weevil Rhynchophorus ferrugineus (Olivier) (Coleoptera: Dryophthoridae) Germ-Free Larvae for Host-Gut Microbes Interaction Studies. BIO-PROTOCOL 2019, 9, e3456. [Google Scholar] [CrossRef]
- Jing, T.-Z.; Qi, F.-H.; Wang, Z.-Y. Most Dominant Roles of Insect Gut Bacteria: Digestion, Detoxification, or Essential Nutrient Provision? Microbiome 2020, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.-H.; Roh, S.W.; Whon, T.W.; Jung, M.-J.; Kim, M.-S.; Park, D.-S.; Yoon, C.; Nam, Y.-D.; Kim, Y.-J.; Choi, J.-H.; et al. Insect Gut Bacterial Diversity Determined by Environmental Habitat, Diet, Developmental Stage, and Phylogeny of Host. Appl. Environ. Microbiol. 2014, 80, 5254–5264. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The Gut Microbiota of Insects—Diversity in Structure and Function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef] [PubMed]
- Farah Nadiah, R.; Norefrina Shafinaz, M.N.; Nurul Wahida, O. Preliminary Study of Gut Bacterial Abundance in Rhynchophorus ferrugineus (Coleoptera: Dryophthoridae) Fed on Different Diets. Serangga 2018, 23, 126–138. [Google Scholar]
- Mason, C.J.; St. Clair, A.; Peiffer, M.; Gomez, E.; Jones, A.G.; Felton, G.W.; Hoover, K. Diet Influences Proliferation and Stability of Gut Bacterial Populations in Herbivorous Lepidopteran Larvae. PLoS ONE 2020, 15, e0229848. [Google Scholar] [CrossRef]
- Liu, Q.-X.; Su, Z.-P.; Liu, H.-H.; Lu, S.-P.; Ma, B.; Zhao, Y.; Hou, Y.-M.; Shi, Z.-H. The Effect of Gut Bacteria on the Physiology of Red Palm Weevil, Rhynchophorus ferrugineus Olivier and Their Potential for the Control of This Pest. Insects 2021, 12, 594. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, A.; Habineza, P.; Ji, T.; Hou, Y.; Shi, Z. Intestinal Microbiota Confer Protection by Priming the Immune System of Red Palm Weevil Rhynchophorus ferrugineus Olivier (Coleoptera: Dryophthoridae). Front. Physiol. 2019, 10, 1303. [Google Scholar] [CrossRef]
- Anbutsu, H.; Moriyama, M.; Nikoh, N.; Hosokawa, T.; Futahashi, R.; Tanahashi, M.; Meng, X.-Y.; Kuriwada, T.; Mori, N.; Oshima, K. Small Genome Symbiont Underlies Cuticle Hardness in Beetles. Proc. Natl. Acad. Sci. USA 2017, 114, E8382–E8391. [Google Scholar] [CrossRef]
- Habineza, P.; Muhammad, A.; Ji, T.; Xiao, R.; Yin, X.; Hou, Y.; Shi, Z. The Promoting Effect of Gut Microbiota on Growth and Development of Red Palm Weevil, Rhynchophorus ferrugineus (Olivier) (Coleoptera: Dryophthoridae) by Modulating Its Nutritional Metabolism. Front. Microbiol. 2019, 10, 1212. [Google Scholar] [CrossRef]
- Manee, M.M.; Alqahtani, F.H.; Al-Shomrani, B.M.; El-Shafie, H.A.F.; Dias, G.B. Omics in the Red Palm Weevil Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae): A Bridge to the Pest. Insects 2023, 14, 255. [Google Scholar] [CrossRef]
- Muhammad, A.; Fang, Y.; Hou, Y.; Shi, Z. The Gut Entomotype of Red Palm Weevil Rhynchophorus ferrugineus Olivier (Coleoptera: Dryophthoridae) and Their Effect on Host Nutrition Metabolism. Front. Microbiol. 2017, 8, 2291. [Google Scholar] [CrossRef]
- Jia, S.; Zhang, X.; Zhang, G.; Yin, A.; Zhang, S.; Li, F.; Wang, L.; Zhao, D.; Yun, Q.; Tala; et al. Seasonally Variable Intestinal Metagenomes of the Red Palm Weevil (Rhynchophorus ferrugineus). Environ. Microbiol. 2013, 15, 3020–3029. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Yang, X.-K.; Zhang, S.-K.; Segraves, K.A.; Xue, H.-J. Parallel Metatranscriptome Analysis Reveals Degradation of Plant Secondary Metabolites by Beetles and Their Gut Symbionts. Mol. Ecol. 2022, 31, 3999–4016. [Google Scholar] [CrossRef]
- Yaman, M.; Ertürk, Ö.; Aslan, İ. Isolation of Some Pathogenic Bacteria from the Great Spruce Bark Beetle, Dendroctonus Micans and Its Specific Predator, Rhizophagus Grandis. Folia Microbiol. 2010, 55, 35–38. [Google Scholar] [CrossRef]
- Anand, A.A.P.; Vennison, S.J.; Sankar, S.G.; Prabhu, D.I.G.; Vasan, P.T.; Raghuraman, T.; Geoffrey, C.J.; Vendan, S.E. Isolation and Characterization of Bacteria from the Gut of Bombyx mori that Degrade Cellulose, Xylan, Pectin and Starch and Their Impact on Digestion. J. Insect Sci. 2010, 10, 107. [Google Scholar] [CrossRef]
- Jancek, S.; Bézier, A.; Gayral, P.; Paillusson, C.; Kaiser, L.; Dupas, S.; Le Ru, B.P.; Barbe, V.; Periquet, G.; Drezen, J.-M.; et al. Adaptive Selection on Bracovirus Genomes Drives the Specialization of Cotesia Parasitoid Wasps. PLoS ONE 2013, 8, e64432. [Google Scholar] [CrossRef] [PubMed]
- Gopinadhan, P.B.; Mohandas, N.; Vasudevan Nair, K.P. Cytoplasmic Polyhedrosis Virus Infecting Redpalm Weevil of Coconut. Curr. Sci. Assoc. 1990, 59, 577–580. [Google Scholar]
- Banerjee, A.; Dangar, T.K. Pseudomonas Aeruginosa, a Facultative Pathogen of Red Palm Weevil, Rhynchophorus ferrugineus. World J. Microbiol. Biotechnol. 1995, 11, 618–620. [Google Scholar] [CrossRef] [PubMed]
- Salama, H.S.; Foda, M.S.; El-Bendary, M.A.; Abdel-Razek, A. Infection of Red Palm Weevil, Rhynchophorus ferrugineus, by Spore-Forming Bacilli Indigenous to Its Natural Habitat in Egypt. J. Pest Sci. 2004, 77, 27–31. [Google Scholar] [CrossRef]
- Muñoz-Benavent, M.; Pérez-Cobas, A.E.; García-Ferris, C.; Moya, A.; Latorre, A. Insects’ Potential: Understanding the Functional Role of Their Gut Microbiome. J. Pharm. Biomed. Anal. 2021, 194, 113787. [Google Scholar] [CrossRef]
- Wang, J.-M.; Bai, J.; Zheng, F.-Y.; Ling, Y.; Li, X.; Wang, J.; Zhi, Y.-C.; Li, X.-J. Diversity of the Gut Microbiome in Three Grasshopper Species Using 16S RRNA and Determination of Cellulose Digestibility. PeerJ 2020, 8, e10194. [Google Scholar] [CrossRef] [PubMed]
- Dantur, K.I.; Enrique, R.; Welin, B.; Castagnaro, A.P. Isolation of Cellulolytic Bacteria from the Intestine of Diatraea Saccharalis Larvae and Evaluation of Their Capacity to Degrade Sugarcane Biomass. AMB Expr. 2015, 5, 15. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, F.; Lu, X. Diversity and Functional Roles of the Gut Microbiota in Lepidopteran Insects. Microorganisms 2022, 10, 1234. [Google Scholar] [CrossRef] [PubMed]
- Juma, G.; Le Ru, B.; Calatayud, P.-A. Assortments of Digestive Enzymes Induced in First Instar Larvae of Busseola Fusca Feeding on Different Plants. Int. J. Insect Sci. 2019, 11, 117954331984352. [Google Scholar] [CrossRef] [PubMed]
- Lwalaba, D.; Hoffmann, K.H.; Woodring, J. Control of the Release of Digestive Enzymes in the Larvae of the Fall Armyworm, Spodoptera frugiperda. Arch. Insect Biochem. Physiol. 2010, 73, 14–29. [Google Scholar] [CrossRef] [PubMed]
- Abd El-latif, A.O. Partial Characterization of the Digestive Proteases and α-Amylase of the Larvae of the Red Palm Weevil, Rhynchophorus ferrugineus. Arthropods 2020, 9, 7–14. [Google Scholar]
- Darrag, H.M.; Almuhanna, H.T.; Hakami, E.H.; Alhojaily, S.M. Analysis of Volatile Secondary Metabolites in Ocimum basilicum Cell Suspensions: Inhibition, In Silico Molecular Docking, and an ADMET Analysis against Proteolytic Enzymes of Rhynchophorus ferrugineus. Plants 2022, 11, 2949. [Google Scholar] [CrossRef]
- Gao, P.; Liu, Z.; Wen, J. Expression Profiling of Plant Cell Wall-Degrading Enzyme Genes in Eucryptorrhynchus scrobiculatus Midgut. Front. Physiol. 2020, 11, 1111. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, M.A.; Ghanem, M.M.E.; Abd-Elaziz, A.M.; Shams-Eldin, I.M. Purification and Characterization of Xylanase Isoenzymes from Red Palm Weevil Rhynchophorus ferrugineus. Biocatal. Agric. Biotechnol. 2018, 14, 321–327. [Google Scholar] [CrossRef]
- Rafiei, V.; Vélëz, H.; Tzelepis, G. The Role of Glycoside Hydrolases in Phytopathogenic Fungi and Oomycetes Virulence. IJMS 2021, 22, 9359. [Google Scholar] [CrossRef]
- Darvishzadeh, A.; Bandani, A.R.; Karimi, J.; Timouri, G. Biochemical Characterisation of Digestive α-Amylase of Red Palm Weevil, Rhynchophorus ferrugineus (Olivier, 1790) (Coleoptera: Curculionidae). Arch. Phytopathol. Plant Prot. 2012, 45, 2132–2142. [Google Scholar] [CrossRef]
- Darvishzadeh, A.; Bandani, A.R. Identification and Enzymatic Characterisation of Digestive Glucosidases from Gut of Red Palm Weevil, Rhynchophorus ferrugineus (Olivier, 1790) (Coleoptera: Curculionidae). Arch. Phytopathol. Plant Prot. 2013, 46, 1159–1167. [Google Scholar] [CrossRef]
- Riseh, N.S.; Ghadamyari, M.; Motamediniya, B. Biochemical Characterisation of α- and β-Glucosidases and α- and β-Galactosidases from Red Palm Weevil, Rhynchophorus ferrugineus (Olivier) (Col.: Curculionide). Plant Prot. Sci. 2012, 48, 85–93. [Google Scholar] [CrossRef]
- Dong, X.; Jiang, Y.; Hur, Y. Genome-Wide Analysis of Glycoside Hydrolase Family 1 β-Glucosidase Genes in Brassica Rapa and Their Potential Role in Pollen Development. IJMS 2019, 20, 1663. [Google Scholar] [CrossRef] [PubMed]
- Laudani, F.; Strano, C.P.; Edwards, M.G.; Malacrinò, A.; Campolo, O.; Halim, H.M.A.E.; Gatehouse, A.M.R.; Palmeri, V. RNAi-Mediated Gene Silencing in Rhynchophorus ferrugineus (Oliver) (Coleoptera: Curculionidae). Open Life Sci. 2017, 12, 214–222. [Google Scholar] [CrossRef]
- Aguilera, C.; Padilla, B.E.; Flórez, C.P.; Rubio, J.D.; Acuña, J.R. RNA interference: Potential uses on functional genomics and genetic control of Hypothenemus hampei (Coleoptera: Scolytinae). Rev. Colomb. Entomol. 2011, 37, 167–172. [Google Scholar] [CrossRef]
- Ghanem, M.M.E.; Mohamed, M.A.; Abd-Elaziz, A.M. Distribution, Purification and Characterization of a Monofunctional Catalase from Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae). Biocatal. Agric. Biotechnol. 2020, 23, 101480. [Google Scholar] [CrossRef]
- Mohamed, M.A.; Ghazy, A.-E.M.; Abdel Karim, G.S.A.; El-khonezy, M.I.; Abd-Elaziz, A.M.; Ghanem, M.M.E. Defense Status in Larval Stage of Red Palm Weevil Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae). Biocatal. Agric. Biotechnol. 2022, 44, 102465. [Google Scholar] [CrossRef]
- Kaur, M.; Chadha, P.; Kaur, S.; Kaur, A. Effect of Aspergillus Flavus on Lipid Peroxidation and Activity of Antioxidant Enzymes in Midgut Tissue of Spodoptera litura Larvae. Arch. Phytopathol. Plant Prot. 2021, 54, 177–190. [Google Scholar] [CrossRef]
- Sandhu, R.K.; Sarao, P.S.; Sharma, N. Antibiosis in Wild Rice Accessions Induced by Nilaparvata lugens (Stål) Feeding. Phytoparasitica 2020, 48, 801–812. [Google Scholar] [CrossRef]
- Kola, V.S.R.; Renuka, P.; Madhav, M.S.; Mangrauthia, S.K. Key Enzymes and Proteins of Crop Insects as Candidate for RNAi Based Gene Silencing. Front. Physiol. 2015, 6, 119. [Google Scholar] [CrossRef] [PubMed]
- Al-Ayedh, H.; Rizwan-ul-Haq, M.; Hussain, A.; Aljabr, A.M. Insecticidal Potency of RNAi-Based Catalase Knockdown in Rhynchophorus ferrugineus (Oliver) (Coleoptera: Curculionidae): Catalase Knockdown in Rhyncophorus ferrugineus. Pest. Manag. Sci. 2016, 72, 2118–2127. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Yi, X.; Hu, Z.; Hu, M.; Chen, S.; Muhammad, R.-H.; Dong, X.; Gong, L. RNAi-Mediated Knockdown of Catalase Causes Cell Cycle Arrest in SL-1 Cells and Results in Low Survival Rate of Spodoptera Litura (Fabricius). PLoS ONE 2013, 8, e59527. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Budachetri, K.; Meyers, V.C.; Karim, S. Assessment of Tick Antioxidant Responses to Exogenous Oxidative Stressors and Insight into the Role of Catalase in the Reproductive Fitness of the Gulf Coast Tick, Amblyomma maculatum: Antioxidant Responses in Gulf Coast Ticks. Insect Mol. Biol. 2016, 25, 283–294. [Google Scholar] [CrossRef]
- Deng, F.; He, Q.; Zhao, Z. Suppressing a Peroxidase Gene Reduces Survival in the Wheat Aphid Sitobion avenae. Arch. Insect Biochem. Physiol. 2016, 93, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fan, J.; Francis, F.; Chen, J. Molecular Characterization and Gene Silencing of Laccase 1 in the Grain Aphid, Sitobion avenae. Arch. Insect Biochem. Physiol. 2018, 97, e21446. [Google Scholar] [CrossRef]
- Das, S.; Radtke, A.; Choi, Y.-J.; Mendes, A.M.; Valenzuela, J.G.; Dimopoulos, G. Transcriptomic and Functional Analysis of the Anopheles Gambiae Salivary Gland in Relation to Blood Feeding. BMC Genom. 2010, 11, 566. [Google Scholar] [CrossRef]
- Wang, M.; Lu, Y.; Cai, Z.; Liang, S.; Niu, Y.; Miao, Y. Phenol Oxidase Is a Necessary Enzyme for the Silkworm Molting Which Is Regulated by Molting Hormone. Mol. Biol. Rep. 2013, 40, 3549–3555. [Google Scholar] [CrossRef]
- Wu, K.; Li, S.; Wang, J.; Ni, Y.; Huang, W.; Liu, Q.; Ling, E. Peptide Hormones in the Insect Midgut. Front. Physiol. 2020, 11, 191. [Google Scholar] [CrossRef]
- Khudri, N.A.F.R.S.; Mohd Masri, M.M.; Maidin, M.S.T.; Kamarudin, N.; Hussain, M.H.; Abd Ghani, I.; Jalinas, J. Preliminary Evaluation of Acoustic Sensors for Early Detection of Red Palm Weevil, Rhynchophorus ferrugineus Incidence on Oil Palm and Coconut in Malaysia. Int. J. Trop. Insect Sci. 2021, 41, 3287–3292. [Google Scholar] [CrossRef]
- AlJabr, A.; Hussain, A.; Rizwan-ul-Haq, M.; Al-Ayedh, H. Toxicity of Plant Secondary Metabolites Modulating Detoxification Genes Expression for Natural Red Palm Weevil Pesticide Development. Molecules 2017, 22, 169. [Google Scholar] [CrossRef]
- Jaffar, S.; Ahmad, S.; Lu, Y. Contribution of Insect Gut Microbiota and Their Associated Enzymes in Insect Physiology and Biodegradation of Pesticides. Front. Microbiol. 2022, 13, 979383. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Rizwan-ul-haq, M.; AlJabr, A.M.; Al-Ayedh, H. Lethality of Sesquiterpenes Reprogramming Red Palm Weevil Detoxification Mechanism for Natural Novel Biopesticide Development. Molecules 2019, 24, 1648. [Google Scholar] [CrossRef] [PubMed]
- Manzoor, M.; Yang, L.; Wu, S.; El-Shafie, H.; Haider, M.S.; Ahmad, J.N. Feeding Preference of Rhynchophorus ferrugineus (Oliver) (Coleoptera: Curculionidae) on Different Date Palm Cultivars and Host Biochemical Responses to Its Infestation. Bull. Entomol. Res. 2022, 112, 494–501. [Google Scholar] [CrossRef]
- Hu, B.; Zhang, S.; Ren, M.; Tian, X.; Wei, Q.; Mburu, D.K.; Su, J. The Expression of Spodoptera exigua P450 and UGT Genes: Tissue Specificity and Response to Insecticides. Insect Sci. 2019, 26, 199–216. [Google Scholar] [CrossRef] [PubMed]
- Pandian, B.A.; Sathishraj, R.; Djanaguiraman, M.; Prasad, P.V.V.; Jugulam, M. Role of Cytochrome P450 Enzymes in Plant Stress Response. Antioxidants 2020, 9, 454. [Google Scholar] [CrossRef]
- Babiker, M.A.A.-B.; Hamadttu, A.F.E.-S. Expression Profiling, Phylogenetic, and Structural Analyses of a Laccase Gene from the Red Palm Weevil, Rhynchophorus ferrugineus. Afr. J. Biotechnol. 2019, 18, 978–990. [Google Scholar] [CrossRef]
- Arakane, Y.; Muthukrishnan, S.; Beeman, R.W.; Kanost, M.R.; Kramer, K.J. Laccase 2 Is the Phenoloxidase Gene Required for Beetle Cuticle Tanning. Proc. Natl. Acad. Sci. USA 2005, 102, 11337–11342. [Google Scholar] [CrossRef]
- Dittmer, N.T.; Gorman, M.J.; Kanost, M.R. Characterization of Endogenous and Recombinant Forms of Laccase-2, a Multicopper Oxidase from the Tobacco Hornworm, Manduca sexta. Insect Biochem. Mol. Biol. 2009, 39, 596–606. [Google Scholar] [CrossRef]
- Ben Thabet, I.; Francis, F.; De Pauw, E.; Besbes, S.; Attia, H.; Deroanne, C.; Blecker, C. Characterisation of Proteins from Date Palm Sap (Phoenix dactylifera L.) by a Proteomic Approach. Food Chem. 2010, 123, 765–770. [Google Scholar] [CrossRef]
- Montesdeoca, M.; Lobo, M.G.; Casanas, N.; Carnero, A.; Castanera, P.; Ortego, F. Partial Characterization of the Proteolytic Enzymes in the Gut of the Banana Weevil, Cosmopolites sordidus, and Effects of Soybean Kunitz Trypsin Inhibitor on Larval Performance. Entomol. Exp. Appl. 2005, 116, 227–236. [Google Scholar] [CrossRef]
- Ranganathan, S.; Ampasala, D.R.; Palaka, B.K.; Ilavarasi, A.V.; Patidar, I.; Poovadan, L.P.; Sapam, T.D. In Silico Binding Profile Analysis and In Vitro Investigation on Chitin Synthase Substrate and Inhibitors from Maize Stem Borer, Chilo partellus. Curr. Comput.-Aided Drug Des. 2021, 17, 881–895. [Google Scholar] [CrossRef]
- Ribeiro, T.P.; Vasquez, D.D.N.; Macedo, L.L.P.; Lourenço-Tessutti, I.T.; Valença, D.C.; Oliveira-Neto, O.B.; Paes-de-Melo, B.; Rodrigues-Silva, P.L.; Firmino, A.A.P.; Basso, M.F.; et al. Stabilized Double-Stranded RNA Strategy Improves Cotton Resistance to CBW (Anthonomus grandis). Int. J. Mol. Sci. 2022, 23, 13713. [Google Scholar] [CrossRef] [PubMed]
- Antony, B.; Johny, J.; Aldosari, S.A.; Abdelazim, M.M. Identification and Expression Profiling of Novel Plant Cell Wall Degrading Enzymes from a Destructive Pest of Palm Trees, Rhynchophorus ferrugineus: Red Palm Weevil Plant Cell Wall Degrading Enzymes. Insect Mol. Biol. 2017, 26, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hameid, N.F. Starvation Effect on Bioactive Components Ofthe Red Palm Weevil Rhynchophorus ferrugineus (Olivier); (Coleoptera: Curculionidae). Sciences 2018, 8, 337–344. [Google Scholar]
- Zhang, H.; Bai, J.; Huang, S.; Liu, H.; Lin, J.; Hou, Y. Neuropeptides and G-Protein Coupled Receptors (GPCRs) in the Red Palm Weevil Rhynchophorus ferrugineus Olivier (Coleoptera: Dryophthoridae). Front. Physiol. 2020, 11, 159. [Google Scholar] [CrossRef]
- Shi, Y.; Pandit, A.; Nachman, R.J.; Christiaens, O.; Davies, S.A.; Dow, J.A.T.; Smagghe, G. Transcriptome Analysis of Neuropeptides in the Beneficial Insect Lacewing (Chrysoperla carnea) Identifies Kinins as a Selective Pesticide Target: A Biostable Kinin Analogue with Activity against the Peach Potato Aphid Myzus persicae. J. Pest Sci. 2023, 96, 253–264. [Google Scholar] [CrossRef]
- Vatanparast, M.; Hosseininaveh, V.; Ghadamyari, M.; Minoo Sajjadian, S. Plant Cell Wall Degrading Enzymes, Pectinase and Cellulase, in the Digestive System of the Red Palm Weevil, Rhynchophorus ferrugineus (Coleoptera: Curculionidae). Plant Prot. Sci. 2014, 50, 190–198. [Google Scholar] [CrossRef]
- Siddiqui, J.A.; Khan, M.M.; Bamisile, B.S.; Hafeez, M.; Qasim, M.; Rasheed, M.T.; Rasheed, M.A.; Ahmad, S.; Shahid, M.I.; Xu, Y. Role of Insect Gut Microbiota in Pesticide Degradation: A Review. Front. Microbiol. 2022, 13, 870462. [Google Scholar] [CrossRef]
- Abdel-Moaty, Z.; Abdelsalam, S. Photosensitizing Effects of Hematoporphyrin Dihydrochloride against the Red Palm Weevil Rhynchophorus ferrugineus (Coleoptera: Curculionidae). Fresenius Environ. Bull. 2021, 30, 8158. [Google Scholar]
- Orfali, R.; Binsuwaileh, A.; Abu Al-Ala’a, H.; Bane-Gamea, S.; Zaidan, N.; Abdelazim, M.; Alhasan Ismael, M.; Perveen, S.; Majrashi, N.; Alluhayb, K.; et al. Production of a Biopesticide on Host and Non-Host Serine Protease Inhibitors for Red Palm Weevil in Palm Trees. Saudi J. Biol. Sci. 2020, 27, 2803–2808. [Google Scholar] [CrossRef]
- Abdelsalam, S.A.; Alzahrani, A.M.; Elmenshawy, O.M.; Abdel-Moneim, A.M. Spinosad Induces Antioxidative Response and Ultrastructure Changes in Males of Red Palm Weevil Rhynchophorus ferrugineus (Coleoptera: Dryophthoridae). J. Insect Sci. 2016, 16, 106. [Google Scholar] [CrossRef] [PubMed]
- Aziza, S.; Mona, E.-D. Toxic and Biochemical Effects of Juniperus communis Essential Oil on the Red Palm Weevil Rhynchophorus ferrugineus (Olivier.) (Coleoptera: Curculionidae). Egypt. J. Biol. Pest Control 2016, 26, 339. [Google Scholar]
- Josephrajkumar, A.; Mohan, C.; Chaturvedi, V.K. Suppression of Growth and Endopeptidases of Red Palm Weevil, Rhynchophorus ferrugineus (Olivier) Infesting Coconut Using Proteinase Inhibitors. Entomon 2016, 41, 283–292. [Google Scholar]
- Riseh, N.S.; Ghadamyari, M. Biochemical Characterization of α-Amylases from Gut and Hemolymph of Rhynchophorus ferrugineus Olivieri (Col.: Curculionidae) and Their Inhibition by Extracts from the Legumes Vigna radiata L. and Phaseolus vulgaris L. Invertebr. Surviv. J. 2012, 9, 72–81. [Google Scholar]
- Lu, Y.; Deng, X.; Zhu, Q.; Wu, D.; Zhong, J.; Wen, L.; Yu, X. The DsRNA Delivery, Targeting and Application in Pest Control. Agronomy 2023, 13, 714. [Google Scholar] [CrossRef]
- Whitten, M.M.A.; Xue, Q.; Taning, C.N.T.; James, R.; Smagghe, G.; Del Sol, R.; Hitchings, M.; Dyson, P. A Narrow Host-Range and Lack of Persistence in Two Non-Target Insect Species of a Bacterial Symbiont Exploited to Deliver Insecticidal RNAi in Western Flower Thrips. Front. Insect Sci. 2023, 3, 1093970. [Google Scholar] [CrossRef]
- Li, H.; Zhong, Q.; Wang, X.; Luo, F.; Zhou, L.; Sun, H.; Yang, M.; Lou, Z.; Chen, Z.; Zhang, X. The Degradation and Metabolism of Chlorfluazuron and Flonicamid in Tea: A Risk Assessment from Tea Garden to Cup. Sci. Total Environ. 2021, 754, 142070. [Google Scholar] [CrossRef]
- Chen, W.; Yang, Q. Development of Novel Pesticides Targeting Insect Chitinases: A Minireview and Perspective. J. Agric. Food Chem. 2020, 68, 4559–4565. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef] [PubMed]


| Enzyme | EC Number | Function |
|---|---|---|
| Trypsin | 3.4.21.4 | Protein digestion |
| Aminopeptidase | 3.4.11.1 | Protein digestion |
| Xylanase | 3.2.1.8 | Xylan (plant cell wall) digestion |
| Glycosidase | 3.2.1 | Hydrolyze polysaccharide of cell wall |
| Amylase | 3.2.1.1 | Starch digestion |
| Catalase | 1.11.1.6 | Catalyze conversion of hydrogen peroxide into oxygen and water |
| Peroxidase | 1.11.1.7 | Catalyze conversion of hydrogen peroxide into oxygen and water |
| Polyphenol oxidase | 1.14.18.1 | Catalyze the oxidation of phenolic compound using oxygen |
| Genes/Transcripts | EC Number | Function |
|---|---|---|
| Cytochrome P450 | * | Catalyze oxidation of xenobiotics |
| glutathione-S-transferase | 2.5.1.18 | Added glutathione to oxidized xenobiotics |
| Laccase | 1.10.3.2 | Cuticle hardening |
| Carboxypeptidase | 3.4.16.2 | Protein digestion |
| Chitin synthase | 2.4.1.16 | Catalyzes the polymerization of chitin polymer |
| Cellulase | 3.2.1.4 | Cell wall degradation (cellulose) |
| Hemicellulase | 3.2.1 | Cell wall degradation (hemicellulose) |
| Pectinase | 3.2.1.15 | Cell wall degradation (pectin) |
| Neuropeptides | - | Trigger physiological process (digestion) |
| Insecticide/Inhibitor | Target Gene/Protein | Function of Target | Reference |
|---|---|---|---|
| Thymus vulgaris and Ocimum basilicum extract; soybean trypsin inhibitor; N-tosyl-l- phenylalanine chloromethyl ketone (chymotrypsin inhibitor) | Trypsin-like serine proteinase assessment; trypsin; chymotrypsin | Protein digestion | [50,51] |
| Hematoporphyrin dihydrochloride (photosensitizer) | Antioxidant enzymes (polyphenol oxidase; peroxidase) | Defense mechanism | [94] |
| Eserine (carbamate inhibitor) | Acetylcholinesterase (AChE) | Detoxifying enzyme | [16] |
| Protease inhibitor from palm dates kernel | Protease | Protein digestion | [95] |
| Novaluron | Chitinase | Chitin regulation | [18] |
| Sesquiterpene (Farnesol, Farnesyl acetate, Picrotoxin); Spinosad | Glutathione S transferase (GST), Cytochrome P450 | Detoxification of xenobiotics | [77,96] |
| Spinosad | Nicotinic acetylcholine receptor and/or gamma Aminobutyric acid (GABA) receptor | Modulation of feeding behavior and reproduction | [96] |
| Juniperus communis essential oil | Gut protein content | Digestion system | [97] |
| RNAi/double strand RNA | Catalase | Defense mechanism | [61,66] |
| Aprotinin | Gut serine proteinase | Serine digestion | [98] |
| Protease inhibitor from Vigna radiata L. seeds | α-amylase | Carbohydrate digestion | [50,99] |
| Not tested | Neuropeptide precursor and receptor | To regulate physiology and behavior of insects | [90] |
| Not tested | Laccase | Oxidize toxic compounds ingested by the insect | [81] |
| Not tested | Xylanase | Digestion of plant cell wall | [53] |
| Not tested | Aminopeptidase | Protein digestion | [20] |
| Not tested | Cellulase | Digestion of plant cell wall | [88] |
| Not tested | Pectinase | Digestion of plant cell walls | [88,92] |
| Not tested | Glucosidase | Carbohydrate digestion | [56] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seman-Kamarulzaman, A.-F.; Pariamiskal, F.A.; Azidi, A.N.; Hassan, M. A Review on Digestive System of Rhynchophorus ferrugineus as Potential Target to Develop Control Strategies. Insects 2023, 14, 506. https://doi.org/10.3390/insects14060506
Seman-Kamarulzaman A-F, Pariamiskal FA, Azidi AN, Hassan M. A Review on Digestive System of Rhynchophorus ferrugineus as Potential Target to Develop Control Strategies. Insects. 2023; 14(6):506. https://doi.org/10.3390/insects14060506
Chicago/Turabian StyleSeman-Kamarulzaman, Ahmad-Faris, Faizatul Atikah Pariamiskal, Amiratul Nabihah Azidi, and Maizom Hassan. 2023. "A Review on Digestive System of Rhynchophorus ferrugineus as Potential Target to Develop Control Strategies" Insects 14, no. 6: 506. https://doi.org/10.3390/insects14060506