Pharmacogenetics of Donepezil and Memantine in Healthy Subjects
Abstract
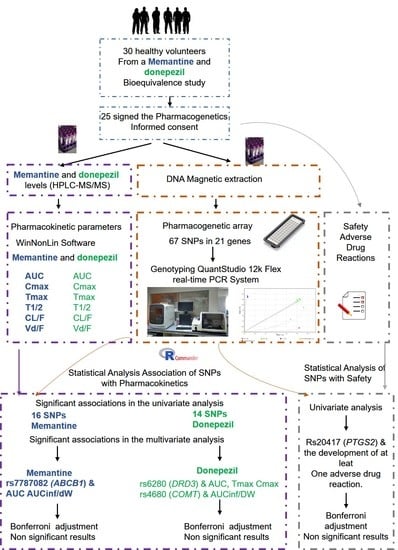
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Study Design and Procedures
2.3. Pharmacokinetic Analysis
2.4. Safety
2.5. Genotyping
2.6. Phenotype Inference
2.7. Statistical Analysis
3. Results
3.1. Demographic Characteristics
3.2. Pharmacokinetics
3.3. Safety
4. Discussion
Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Owen, R.T. Memantine and Donepezil: A Fixed Drug Combination for the Treatment of Moderate to Severe Alzheimer’s Dementia. Drugs Today Barc. Spain 1998 2016, 52, 239–248. [Google Scholar] [CrossRef]
- Takahashi-Ito, K.; Makino, M.; Okado, K.; Tomita, T. Memantine Inhibits β-Amyloid Aggregation and Disassembles Preformed β-Amyloid Aggregates. Biochem. Biophys. Res. Commun. 2017, 493, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Reddy, P.H. Role of Glutamate and NMDA Receptors in Alzheimer’s Disease. J. Alzheimers Dis. JAD 2017, 57, 1041–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parsons, C.G.; Danysz, W.; Dekundy, A.; Pulte, I. Memantine and Cholinesterase Inhibitors: Complementary Mechanisms in the Treatment of Alzheimer’s Disease. Neurotox. Res. 2013, 24, 358–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, X.; Jensen, M.Ø.; Jogini, V.; Stein, R.A.; Lee, C.-H.; Mchaourab, H.S.; Shaw, D.E.; Gouaux, E. Mechanism of NMDA Receptor Channel Block by MK-801 and Memantine. Nature 2018, 556, 515–519. [Google Scholar] [CrossRef]
- McKeage, K. Memantine: A Review of Its Use in Moderate to Severe Alzheimer’s Disease. CNS Drugs 2009, 23, 881–897. [Google Scholar] [CrossRef]
- European Medicines Agency Summary of Products Characteristitcs Ebixa. 2002. Available online: https://www.emaeuropaeuendocumentsproduct-Informationebixa-Epar-Prod.-Informationen.pdf (accessed on 5 May 2021).
- Zúñiga Santamaría, T.; Yescas Gómez, P.; Fricke Galindo, I.; González González, M.; Ortega Vázquez, A.; López López, M. Estudios farmacogenéticos en la enfermedad de Alzheimer. Neurología 2018, 37, 287–303. [Google Scholar] [CrossRef]
- European Medicines Agency Aricept: Summary of Product Characteristics; EMC: Hatfield, UK, 1998.
- Birks, J.S.; Harvey, R.J. Donepezil for Dementia Due to Alzheimer’s Disease. Cochrane Database Syst. Rev. 2018, 2018, CD001190. [Google Scholar] [CrossRef]
- Rogers, S.L.; Friedhoff, L.T. Pharmacokinetic and Pharmacodynamic Profile of Donepezil HCl Following Single Oral Doses. Br. J. Clin. Pharmacol. 1998, 46 (Suppl. 1), 1–6. [Google Scholar] [CrossRef] [Green Version]
- Seripa, D.; Bizzarro, A.; Pilotto, A.; D’onofrio, G.; Vecchione, G.; Gallo, A.P.; Cascavilla, L.; Paris, F.; Grandone, E.; Mecocci, P.; et al. Role of Cytochrome P4502D6 Functional Polymorphisms in the Efficacy of Donepezil in Patients with Alzheimer’s Disease. Pharmacogenet. Genom. 2011, 21, 225–230. [Google Scholar] [CrossRef]
- Tiseo, P.J.; Perdomo, C.A.; Friedhoff, L.T. Metabolism and Elimination of 14C-Donepezil in Healthy Volunteers: A Single-Dose Study. Br. J. Clin. Pharmacol. 1998, 46 (Suppl. 1), 19–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noetzli, M.; Guidi, M.; Ebbing, K.; Eyer, S.; Wilhelm, L.; Michon, A.; Thomazic, V.; Stancu, I.; Alnawaqil, A.-M.; Bula, C.; et al. Population Pharmacokinetic Approach to Evaluate the Effect of CYP2D6, CYP3A, ABCB1, POR and NR1I2 Genotypes on Donepezil Clearance. Br. J. Clin. Pharmacol. 2014, 78, 135–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.-H.; Wu, S.-L.; Chou, M.-C.; Lai, C.-L.; Chen, S.-H.; Liu, C.-K. Plasma Concentration of Donepezil to the Therapeutic Response of Alzheimer’s Disease in Taiwanese. J. Alzheimers Dis. 2011, 23, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Noetzli, M.; Eap, C.B. Pharmacodynamic, Pharmacokinetic and Pharmacogenetic Aspects of Drugs Used in the Treatment of Alzheimer’s Disease. Clin. Pharmacokinet. 2013, 52, 225–241. [Google Scholar] [CrossRef] [PubMed]
- Roden, D.M.; McLeod, H.L.; Relling, M.V.; Williams, M.S.; Mensah, G.A.; Peterson, J.F.; Van Driest, S.L. Pharmacogenomics. Lancet 2019, 394, 521–532. [Google Scholar] [CrossRef]
- Blesa, R.; Bullock, R.; He, Y.; Bergman, H.; Gambina, G.; Meyer, J.; Rapatz, G.; Nagel, J.; Lane, R. Effect of Butyrylcholinesterase Genotype on the Response to Rivastigmine or Donepezil in Younger Patients with Alzheimer’s Disease. Pharmacogenet. Genom. 2006, 16, 771–774. [Google Scholar] [CrossRef] [Green Version]
- Sokolow, S.; Li, X.; Chen, L.; Taylor, K.D.; Rotter, J.I.; Rissman, R.A.; Aisen, P.S.; Apostolova, L.G. Deleterious Effect of Butyrylcholinesterase K-Variant in Donepezil Treatment of Mild Cognitive Impairment. J. Alzheimer’s Dis. JAD 2017, 56, 229–237. [Google Scholar] [CrossRef] [Green Version]
- Cacabelos, R. Donepezil in Alzheimer’s Disease: From Conventional Trials to Pharmacogenetics. Neuropsychiatr. Dis. Treat. 2007, 3, 303–333. [Google Scholar]
- Pilotto, A.; Franceschi, M.; D’Onofrio, G.; Bizzarro, A.; Mangialasche, F.; Cascavilla, L.; Paris, F.; Matera, M.G.; Pilotto, A.; Daniele, A.; et al. Effect of a CYP2D6 Polymorphism on the Efficacy of Donepezil in Patients with Alzheimer Disease. Neurology 2009, 73, 761–767. [Google Scholar] [CrossRef] [Green Version]
- Klimkowicz-Mrowiec, A.; Wolkow, P.; Sado, M.; Dziubek, A.; Pera, J.; Dziedzic, T.; Szczudlik, A.; Slowik, A. Influence of Rs1080985 Single Nucleotide Polymorphism of the CYP2D6 Gene on Response to Treatment with Donepezil in Patients with Alzheimer’s Disease. Neuropsychiatr. Dis. Treat. 2013, 9, 1029–1033. [Google Scholar] [CrossRef] [Green Version]
- Ortner, M.; Stange, M.; Schneider, H.; Schröder, C.; Buerger, K.; Müller, C.; Müller-Sarnowski, F.; Diehl-Schmid, J.; Förstl, H.; Grimmer, T.; et al. Therapeutic Drug Monitoring of Rivastigmine and Donepezil Under Consideration of CYP2D6 Genotype-Dependent Metabolism of Donepezil. Drug Des. Dev. Ther. 2020, 14, 3251–3262. [Google Scholar] [CrossRef]
- Lu, J.; Wang, X.; Wan, L.; Fu, J.; Huo, Y.; Zhao, Y.; Guo, C. Gene Polymorphisms Affecting the Pharmacokinetics and Pharmacodynamics of Donepezil Efficacy. Front. Pharmacol. 2020, 11, 934. [Google Scholar] [CrossRef] [PubMed]
- European Medicines Agency Summary of Products Characteristics: Donepezil Hydrocloride. Available online: https://cima.aemps.es/cima/pdfs/es/ft/75249/75249_ft.pdf (accessed on 5 May 2021).
- Noetzli, M.; Guidi, M.; Ebbing, K.; Eyer, S.; Wilhelm, L.; Michon, A.; Thomazic, V.; Alnawaqil, A.-M.; Maurer, S.; Zumbach, S.; et al. Population Pharmacokinetic Study of Memantine: Effects of Clinical and Genetic Factors. Clin. Pharmacokinet. 2013, 52, 211–223. [Google Scholar] [CrossRef]
- Aguirre, C.; García, M. Causality assessment in reports on adverse drug reactions. Algorithm of Spanish pharmacovigilance system. Med. Clin. 2016, 147, 461–464. [Google Scholar] [CrossRef]
- Saiz-Rodríguez, M.; Belmonte, C.; Román, M.; Ochoa, D.; Koller, D.; Talegón, M.; Ovejero-Benito, M.C.; López-Rodríguez, R.; Cabaleiro, T.; Abad-Santos, F. Effect of Polymorphisms on the Pharmacokinetics, Pharmacodynamics and Safety of Sertraline in Healthy Volunteers. Basic Clin. Pharmacol. Toxicol. 2018, 122, 501–511. [Google Scholar] [CrossRef]
- Desta, Z.; Gammal, R.S.; Gong, L.; Whirl-Carrillo, M.; Gaur, A.H.; Sukasem, C.; Hockings, J.; Myers, A.; Swart, M.; Tyndale, R.F.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2B6 and Efavirenz-Containing Antiretroviral Therapy. Clin. Pharmacol. Ther. 2019, 106, 726–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caudle, K.E.; Sangkuhl, K.; Whirl-Carrillo, M.; Swen, J.J.; Haidar, C.E.; Klein, T.E.; Gammal, R.S.; Relling, M.V.; Scott, S.A.; Hertz, D.L.; et al. Standardizing CYP2D6 Genotype to Phenotype Translation: Consensus Recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin. Transl. Sci. 2020, 13, 116–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, S.A.; Sangkuhl, K.; Stein, C.M.; Hulot, J.-S.; Mega, J.L.; Roden, D.M.; Klein, T.E.; Sabatine, M.S.; Johnson, J.A.; Shuldiner, A.R. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C19 Genotype and Clopidogrel Therapy: 2013 Update. Clin. Pharmacol. Ther. 2013, 94, 317–323. [Google Scholar] [CrossRef]
- Caudle, K.E.; Rettie, A.E.; Whirl-Carrillo, M.; Smith, L.H.; Mintzer, S.; Lee, M.T.M.; Klein, T.E.; Callaghan, J.T. Clinical Pharmacogenetics Implementation Consortium Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and HLA-B Genotypes and Phenytoin Dosing. Clin. Pharmacol. Ther. 2014, 96, 542–548. [Google Scholar] [CrossRef] [Green Version]
- Saiz-Rodríguez, M.; Ochoa, D.; Belmonte, C.; Román, M.; Vieira de Lara, D.; Zubiaur, P.; Koller, D.; Mejía, G.; Abad-Santos, F. Polymorphisms in CYP1A2, CYP2C9 and ABCB1 Affect Agomelatine Pharmacokinetics. J. Psychopharmacol. Oxf. Engl. 2019, 33, 522–531. [Google Scholar] [CrossRef] [Green Version]
- Saiz-Rodríguez, M.; Belmonte, C.; Caniego, J.L.; Koller, D.; Zubiaur, P.; Bárcena, E.; Romero-Palacián, D.; Eugene, A.R.; Ochoa, D.; Abad-Santos, F. Influence of CYP450 Enzymes, CES1, PON1, ABCB1, and P2RY12 Polymorphisms on Clopidogrel Response in Patients Subjected to a Percutaneous Neurointervention. Clin. Ther. 2019, 41, 1199–1212.e2. [Google Scholar] [CrossRef] [PubMed]
- Sole, X.; Guino, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A Web Tool for the Analysis of Association Studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, N.; Rodrigues, C.; Lamba, M.; Wu, W.; Bronskill, S.E.; Herrmann, N.; Gill, S.S.; Chan, A.-W.; Mason, R.; Day, S.; et al. Systematic Review of Sex-Specific Reporting of Data: Cholinesterase Inhibitor Example. J. Am. Geriatr. Soc. 2017, 65, 2213–2219. [Google Scholar] [CrossRef] [PubMed]
- Giacobini, E.; Pepeu, G. Sex and Gender Differences in the Brain Cholinergic System and in the Response to Therapy of Alzheimer Disease with Cholinesterase Inhibitors. Curr. Alzheimer Res. 2018, 15, 1077–1084. [Google Scholar] [CrossRef] [PubMed]
- Scacchi, R.; Gambina, G.; Broggio, E.; Corbo, R.M. Sex and ESR1 Genotype May Influence the Response to Treatment with Donepezil and Rivastigmine in Patients with Alzheimer’s Disease. Int. J. Geriatr. Psychiatry 2014, 29, 610–615. [Google Scholar] [CrossRef] [PubMed]
- Canevelli, M.; Quarata, F.; Remiddi, F.; Lucchini, F.; Lacorte, E.; Vanacore, N.; Bruno, G.; Cesari, M. Sex and Gender Differences in the Treatment of Alzheimer’s Disease: A Systematic Review of Randomized Controlled Trials. Pharmacol. Res. 2017, 115, 218–223. [Google Scholar] [CrossRef]
- Van Marum, R.J. Update on the Use of Memantine in Alzheimer’s Disease. Neuropsychiatr. Dis. Treat. 2009, 5, 237–247. [Google Scholar] [CrossRef] [Green Version]
- Kagawa, Y.; Yamamoto, Y.; Ueno, A.; Maeda, T.; Obi, T. Impact of CYP2D6, CYP3A5, and ABCB1 Polymorphisms on Plasma Concentrations of Donepezil and Its Metabolite in Patients with Alzheimer’s Disease. Ther. Drug Monit. 2020, 43, 429–435. [Google Scholar] [CrossRef]
- Proitsi, P.; Powell, J.F. Missense Substitutions Associated with Behavioural Disturbances in Alzheimer’s Disease (AD). Brain Res. Bull. 2012, 88, 394–405. [Google Scholar] [CrossRef]
- Patel, C.N.; Georrge, J.J.; Modi, K.M.; Narechania, M.B.; Patel, D.P.; Gonzalez, F.J.; Pandya, H.A. Pharmacophore-Based Virtual Screening of Catechol-o-Methyltransferase (COMT) Inhibitors to Combat Alzheimer’s Disease. J. Biomol. Struct. Dyn. 2018, 36, 3938–3957. [Google Scholar] [CrossRef]
- Yoon, H.; Myung, W.; Lim, S.-W.; Kang, H.S.; Kim, S.; Won, H.-H.; Carroll, B.J.; Kim, D.K. Association of the Choline Acetyltransferase Gene with Responsiveness to Acetylcholinesterase Inhibitors in Alzheimer’s Disease. Pharmacopsychiatry 2015, 48, 111–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scacchi, R.; Gambina, G.; Moretto, G.; Corbo, R.M. Variability of AChE, BChE, and ChAT Genes in the Late-Onset Form of Alzheimer’s Disease and Relationships with Response to Treatment with Donepezil and Rivastigmine. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. Off. Publ. Int. Soc. Psychiatr. Genet. 2009, 150B, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Braga, I.L.S.; Silva, P.N.; Furuya, T.K.; Santos, L.C.; Pires, B.C.; Mazzotti, D.R.; Bertolucci, P.H.; Cendoroglo, M.S.; Smith, M.C. Effect of APOE and CHRNA7 Genotypes on the Cognitive Response to Cholinesterase Inhibitor Treatment at Different Stages of Alzheimer’s Disease. Am. J. Alzheimer’s Dis. Other Demen. 2015, 30, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Sonkusare, S.K.; Kaul, C.L.; Ramarao, P. Dementia of Alzheimer’s Disease and Other Neurodegenerative Disorders--Memantine, a New Hope. Pharmacol. Res. 2005, 51, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, N.; Li, A.; Lanctôt, K. Memantine in Dementia: A Review of the Current Evidence. Expert Opin. Pharmacother. 2011, 12, 787–800. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Liang, B.; Wang, Z.; Cheng, X.; Huang, Y.; Liu, Y.; Huang, Z. Influence of Four Polymorphisms in ABCA1 and PTGS2 Genes on Risk of Alzheimer’s Disease: A Meta-Analysis. Neurol. Sci. Off. J. Ital. Neurol. Soc. Ital. Soc. Clin. Neurophysiol. 2016, 37, 1209–1220. [Google Scholar] [CrossRef]
- Rammes, G.; Rupprecht, R.; Ferrari, U.; Zieglgänsberger, W.; Parsons, C.G. The N-Methyl-D-Aspartate Receptor Channel Blockers Memantine, MRZ 2/579 and Other Amino-Alkyl-Cyclohexanes Antagonise 5-HT(3) Receptor Currents in Cultured HEK-293 and N1E-115 Cell Systems in a Non-Competitive Manner. Neurosci. Lett. 2001, 306, 81–84. [Google Scholar] [CrossRef]
- Seeman, P.; Caruso, C.; Lasaga, M. Memantine Agonist Action at Dopamine D2High Receptors. Synapse 2008, 62, 149–153. [Google Scholar] [CrossRef]
- Tanila, H. The Role of BDNF in Alzheimer’s Disease. Neurobiol. Dis. 2017, 97, 114–118. [Google Scholar] [CrossRef] [Green Version]
- Leyhe, T.; Stransky, E.; Eschweiler, G.W.; Buchkremer, G.; Laske, C. Increase of BDNF Serum Concentration during Donepezil Treatment of Patients with Early Alzheimer’s Disease. Eur. Arch. Psychiatry Clin. Neurosci. 2008, 258, 124–128. [Google Scholar] [CrossRef]
- Micuda, S.; Mundlova, L.; Anzenbacherova, E.; Anzenbacher, P.; Chladek, J.; Fuksa, L.; Martinkova, J. Inhibitory Effects of Memantine on Human Cytochrome P450 Activities: Prediction of in Vivo Drug Interactions. Eur. J. Clin. Pharmacol. 2004, 60, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Tatton, W.; Chen, D.; Chalmers-Redman, R.; Wheeler, L.; Nixon, R.; Tatton, N. Hypothesis for a Common Basis for Neuroprotection in Glaucoma and Alzheimer’s Disease: Anti-Apoptosis by Alpha-2-Adrenergic Receptor Activation. Surv. Ophthalmol. 2003, 48 (Suppl. 1), S25–S37. [Google Scholar] [CrossRef]
- Cacabelos, R.; Meyyazhagan, A.; Carril, J.C.; Cacabelos, P.; Teijido, Ó. Pharmacogenetics of Vascular Risk Factors in Alzheimer’s Disease. J. Pers. Med. 2018, 8, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cacabelos, R. Pharmacogenomic Protocols in CNS Disorders and Dementia. Neurodegener. Dis. 2010, 7, 167–169. [Google Scholar] [CrossRef]
- Zhou, S.-F.; Zhou, Z.-W.; Yang, L.-P.; Cai, J.-P. Substrates, Inducers, Inhibitors and Structure-Activity Relationships of Human Cytochrome P450 2C9 and Implications in Drug Development. Curr. Med. Chem. 2009, 16, 3480–3675. [Google Scholar] [CrossRef]
- Craig, D.; Hart, D.J.; Carson, R.; McIlroy, S.P.; Passmore, A.P. Psychotic Symptoms in Alzheimer’s Disease Are Not Influenced by Polymorphic Variation at the Dopamine Receptor DRD3 Gene. Neurosci. Lett. 2004, 368, 33–36. [Google Scholar] [CrossRef]
- Kodani, S.D.; Morisseau, C. Role of Epoxy-Fatty Acids and Epoxide Hydrolases in the Pathology of Neuro-Inflammation. Biochimie 2019, 159, 59–65. [Google Scholar] [CrossRef]
- Hayslett, R.L.; Tizabi, Y. Effects of Donepezil, Nicotine and Haloperidol on the Central Serotonergic System in Mice: Implications for Tourette’s Syndrome. Pharmacol. Biochem. Behav. 2005, 81, 879–886. [Google Scholar] [CrossRef]
| Demographic Feature | Total (n = 25) | Females (n = 12) | Males (n = 13) | p-Value | |
|---|---|---|---|---|---|
| Age (years) | 30.64 ± 7.64 | 33.5 ± 9.62 | 28.00 ± 4.02 | 0.087 | |
| Ethnic (%) | Latin | 19 (76.0%) | 8 (66.7%) | 11 (84.6%) | 0.378 |
| Caucasian | 6 (24.0%) | 4 (33.3%) | 2 (15.4%) | ||
| Height (cm) | 167.48 ± 10.51 | 158.42 ± 4.17 | 175.85 ± 6.87 | 0.000 * | |
| Weight (kg) | 69.58 ± 12.87 | 59.86 ± 7.74 | 78.56 ± 9.73 | 0.000 * | |
| BMI (kg/m2) | 24.63 ± 2.65 | 23.85 ± 2.87 | 25.36 ± 2.3 | 0.160 | |
| DW (mg/kg) | 0.15 ± 0.03 | 0.17 ± 0.02 | 0.13 ± 0.02 | 0.000 * | |
| Drug | Pharmacokinetic Parameter | Females (n = 12) | Males (n = 7) | Total (n = 16) | p-Value |
|---|---|---|---|---|---|
| Donepezil | AUC (ng∗h/mL) | 734.55 ± 229.74 | 576.33 ± 159.74 | 652.28 ± 208.47 | 0.058 |
| AUC/DW (ng∗h∗mg/mL × kg) | 4301.37 ± 1169.75 | 4444.19 ± 976.87 | 4375.64 ± 1053.37 | 0.684 | |
| Tmax (h) | 2.23 ± 0.76 | 2.37 ± 0.66 | 2.30 ± 0.70 | 0.544 | |
| Cmax (ng/mL) | 20.49 ± 5.22 | 17.00 ± 4.48 | 18.67 ± 5.07 | 0.087 | |
| Cmax/DW (ng∗mg/mL∗kg) | 120.73 ± 25.91 | 130.49 ± 24.72 | 125.81 ± 25.26 | 0.332 | |
| Vd/F (L/kg) | 15.87 ± 3.19 | 13.17 ± 2.20 | 14.44 ± 3.01 | 0.015 * | |
| Cl/F (L/h × kg) | 0.25 ± 0.06 | 0.24 ± 0.06 | 0.24 ± 0.05 | 0.704 | |
| T½ (h) | 46.139 ± 9.96 | 40.34 ± 10.51 | 43.112 ± 10.46 | 0.139 | |
| Memantine | AUC (ng × h/mL) | 280.42 ± 0.28 | 153.52 ± 0.154 | 263.92 ± 0.26 | 0.005 * |
| AUC/DW (ng × h × mg/mL × kg) | 1550.07 ± 1.55 | 969.10 ± 0.97 | 1253.68 ± 1.25 | 0.966 | |
| Tmax (h) | 5.94 ± 2.26 | 5.12 ± 2.11 | 5.51 ± 2.18 | 0.362 | |
| Cmax (ng/mL) | 18.07 ± 3.06 | 13.37 ± 2.28 | 15.63 ± 3.55 | 0.000 * | |
| Cmax/DW (ng × mg/mL × kg) | 107.027 ± 15.67 | 103.33 ± 9.39 | 105.10 ± 12.66 | 0.570 | |
| Vd/F (L/kg) | 9.53 ± 1.22 | 9.59 ± 0.91 | 9.56 ± 1.05 | 0.846 | |
| Cl/F (L/h × kg) | 0.15 ± 0.03 | 0.15 ± 0.022 | 0.15 ± 0.03 | 0.936 | |
| T½ (h) | 45.56 ± 10.28 | 45.67 ± 8.02 | 45.61 ± 8.98 | 0.882 |
| Genotype/Phenotype | N | AUC (ng∗h/mL) | AUC/dW (ng∗h∗mg/mL∗kg) | Tmax (h) | Cmax (pg/mL) | Cmax/dW (ng∗mg/mL∗kg) | T½ (h) | Vd/F (L/kg) | Cl/F (L/h∗kg) | |
|---|---|---|---|---|---|---|---|---|---|---|
| ABCB1 rs10248420 | G/G | 1 | 822.74 ± 0 | 7314.18 ± 0 | 7.25 ± 0 | 10.79 ± 0.00 * | 95.9 ± 0 | 48.39 ± 0 | 9.54 ± 0 | 0.14 ± 0 |
| A/G | 12 | 1138.78 ± 282.22 | 7016.85 ± 1584.48 | 6.17 ± 2.15 | 17.1 ± 2.7 | 105.9 ± 15.99 | 45.63 ± 10.36 | 9.45 ± 1.27 | 0.15 ± 0.03 | |
| A/A | 12 | 926.16 ± 209.36 | 6747.51 ± 928.86 | 4.71 ± 2.07 | 14.56 ± 3.81 | 105.07 ± 9.23 | 45.37 ± 8.23 | 9.68 ± 0.87 | 0.15 ± 0.02 | |
| ABCB1 rs10276036 | T/T | 9 | 1138.79 ± 306.45 | 7165.01 ± 1717.57 | 6.72 ± 2.06 * | 16.43 ± 3.2 | 103.26 ± 15.02 | 47.18 ± 10.91 | 9.56 ± 1.21 | 0.15 ± 0.03 |
| T/C | 16 | 959.55 ± 221.42 | 6750.09 ± 935.56 | 4.83 ± 1.98 | 15.18 ± 3.76 | 106.14 ± 11.52 | 44.73 ± 7.95 | 9.56 ± 0.99 | 0.15 ± 0.02 | |
| ABCB1 rs7787082 | A/A | 2 | 781.52 ± 58.3 * | 6111.17 ± 1701.31 | 6.13 ± 1.59 | 12.18 ± 1.97 *,#1 | 92.92 ± 4.21 | 41.78 ± 9.34 | 9.93 ± 0.55 | 0.17 ± 0.05 |
| A/G | 11 | 1175 ± 265.12 | 7208.55 ± 1508.82 | 6.27 ± 2.22 | 17.42 ± 2.59 | 107.35 ± 15.92 | 46.58 ± 10.3 | 9.37 ± 1.3 | 0.14 ± 0.03 | |
| G/G | 12 | 926.16 ± 209.36 | 6747.51 ± 928.86 | 4.71 ± 2.07 | 14.56 ± 3.81 | 105.07 ± 9.23 | 45.37 ± 8.23 | 9.68 ± 0.87 | 0.15 ± 0.02 | |
| ADRA2A rs1800544 | C/C | 7 | 948.63 ± 219.69 | 6517.46 ± 978.66 | 5.39 ± 2.35 | 15.49 ± 4.07 | 105.06 ± 8.47 | 42.6 ± 7.95 * | 9.4 ± 0.66 | 0.16 ± 0.02 |
| G/C | 15 | 989.44 ± 249.17 | 6808.31 ± 1122.71 | 4.97 ± 1.8 | 15.51 ± 3.67 | 106.28 ± 12.86 | 44.61 ± 8.5 | 9.47 ± 1.04 | 0.15 ± 0.03 | |
| G/G | 3 | 1373.34 ± 211.15 | 8246.54 ± 1966.97 | 8.5 ± 1.32 | 16.57 ± 2.52 | 99.32 ± 22.1 #2 | 57.68 ± 3.75 | 10.37 ± 1.78 | 0.13 ± 0.03 | |
| APOC3 rs4520 | C/C | 16 | 1045.44 ± 246.52 | 7110.71 ± 1270.46 | 6.17 ± 2.12 * | 15.06 ± 2.87 | 102.38 ± 11.15 | 47.91 ± 9.2 | 9.74 ± 1.04 | 0.15 ± 0.02 |
| C/T | 9 | 986.11 ± 304.11 | 6523.89 ± 1200.34 | 4.33 ± 1.82 | 16.64 ± 4.54 | 109.95 ± 14.35 | 41.53 ± 7.33 | 9.25 ± 1.05 | 0.16 ± 0.03 | |
| CYP2C9 | NM | 21 | 1009.08 ± 280.65 | 6984.66 ± 1341.13 | 5.71 ± 2.22 | 14.84 ± 3.17 * | 103.03 ± 11.86 | 46.94 ± 9.03 | 9.73 ± 0.98 * | 0.15 ± 0.03 |
| IM | 4 | 1102.82 ± 149.36 | 6452.13 ± 515.02 | 4.44 ± 1.8 | 19.78 ± 2.59 | 115.97 ± 12.5 | 38.64 ± 4.95 | 8.67 ± 1.1 | 0.16 ± 0.01 | |
| CYP2D6 | UM + NM | 2 | 976.08 ± 243.70 * | 6723.09 ± 1053.94∗ | 5.43 ± 2.31 | 15.15 ± 3.56 | 104.24 ± 12.98 | 44.70 ± 8.68 | 9.60 ± 1.10 | 0.15 ± 0.02 |
| IM+ PM | 2 | 1276.10 ± 246.00 * | 7825.39 ± 1951.29 * | 5.94 ± 1.46 | 18.12 ± 2.60 | 109.62 ± 11.21 | 50.44 ± 10.29 | 9.38 ± 0.86 | 0.13 ± 0.03 | |
| HTR2A rs6314 | C/C | 21 | 1012.16 ± 285.94 | 6751.67 ± 1277.64 | 5.61 ± 1.95 | 15.99 ± 3.72 | 106.59 ± 11.25 | 43.57 ± 8.12 * | 9.35 ± 0.86 * | 0.15 ± 0.03 |
| C/T | 4 | 1086.67 ± 77.14 | 7675.35 ± 857.71 | 5 ± 3.47 | 13.71 ± 1.71 | 97.29 ± 18.45 | 56.33 ± 4.79 | 10.69 ± 1.37 | 0.13 ± 0.02 | |
| HTR2C rs518147 | C/C | 11 | 839.26 ± 152.51 *,#3 | 6550.63 ± 919.57 | 5.14 ± 2.22 | 13.38 ± 2.32 *,#3 | 103.84 ± 8.3 | 42.93 ± 6.49 | 9.47 ± 0.76 | 0.16 ± 0.02 |
| G/C | 6 | 1165.53 ± 306.44 | 6843.25 ± 1843.45 | 6.29 ± 1.61 | 18.01 ± 3.11 | 105.38 ± 11.4 | 44.82 ± 10.17 | 9.55 ± 0.88 | 0.15 ± 0.04 | |
| G/G | 8 | 1172.12 ± 206.5 #4 | 7421.25 ± 1117.26 | 5.44 ± 2.57 | 16.94 ± 3.73 | 106.62 ± 18.77 | 49.9 ± 10.46 | 9.7 ± 1.54 | 0.14 ± 0.02 | |
| Total | 25 | 1024.08 ± 263.92 | 6899.46 ± 1253.68 | 5.51 ± 2.18 | 15.63 ± 3.55 | 105.1 ± 12.65 | 45.61 ± 8.98 | 9.56 ± 1.05 | 0.15 ± 0.03 |
| Genotype/Phenotype | N | AUC (ng∗h/mL) | AUC/dW (ng∗h∗mg/mL∗kg) | Tmax (h) | Cmax (pg/mL) | Cmax/dW (ng ×*mg/mL∗kg) | T½ (h) | Vd/F (L/kg) | Cl/F (L/h∗kg) | |
|---|---|---|---|---|---|---|---|---|---|---|
| ABCB1 rs10280101 | A/A | 16 | 668.75 ± 242.41 | 4573.19 ± 1159.48 | 2.36 ± 0.7 | 18.44 ± 5.06 | 128.1 ± 24.7 | 42.55 ± 12.21 | 13.4 ± 1.91 * | 0.23 ± 0.06 |
| A/C | 9 | 622.99 ± 136.71 | 4024.44 ± 768.91 | 2.19 ± 0.73 | 19.08 ± 5.35 | 121.74 ± 27.22 | 44.13 ± 6.86 | 16.29 ± 3.78 * | 0.26 ± 0.05 | |
| ABCB1 rs11983225 | T/T | 16 | 668.75 ± 242.41 | 4573.19 ± 1159.48 | 2.36 ± 0.7 | 18.44 ± 5.06 | 128.1 ± 24.7 | 42.55 ± 12.21 | 13.4 ± 1.91 * | 0.23 ± 0.06 |
| T/C | 9 | 622.99 ± 136.71 | 4024.44 ± 768.91 | 2.19 ± 0.73 | 19.08 ± 5.35 | 121.74 ± 27.22 | 44.13 ± 6.86 | 16.29 ± 3.78 * | 0.26 ± 0.05 | |
| ABCB1 rs3842 | T/T | 20 | 670.65 ± 223.03 | 4446.73 ± 1117.81 | 2.13 ± 0.60 * | 19.69 ± 4.78 * | 131.48 ± 21.66 * | 42.61 ± 11.24 | 13.97 ± 2.57 | 0.24 ± 0.06 |
| T/C | 5 | 578.77 ± 126.59 | 4091.26 ± 772.05 | 3 ± 0.68 * | 14.6 ± 4.41 * | 103.15 ± 28.23 * | 45.15 ± 7.04 | 16.31 ± 4.19 | 0.25 ± 0.05 | |
| ABCB1 rs4728709 | A/G | 8 | 638.47 ± 255.84 | 4450.28 ± 1250.16 | 2.69 ± 0.58 * | 16.48 ± 4.04 | 117.5 ± 23.82 | 43.32 ± 13.48 | 13.97 ± 1.93 | 0.24 ± 0.05 |
| G/G | 17 | 658.77 ± 190.82 | 4340.51 ± 988.18 | 2.12 ± 0.69 * | 19.7 ± 5.28 | 129.72 ± 25.65 | 43.02 ± 9.19 | 14.66 ± 3.43 | 0.24 ± 0.06 | |
| ABCB1 rs7787082 | A/A | 2 | 485.06 ± 58.65 | 3810.89 ± 1230.23 | 2.25 ± 1.06 | 12.01 ± 2.85 *,#1 | 90.92 ± 2.9 | 46.59 ± 8.47 | 17.98 ± 2.67 | 0.28 ± 0.09 |
| A/G | 11 | 710.3 ± 198.75 | 4354.73 ± 1164.04 | 2.32 ± 0.74 | 20.18 ± 3.97 * | 124.64 ± 25.56 | 44.35 ± 11.08 | 15 ± 3.39 | 0.24 ± 0.05 | |
| G/G | 12 | 626.96 ± 222.36 | 4488.92 ± 989.49 | 2.29 ± 0.68 | 18.4 ± 5.52 * | 132.7 ± 22.86 | 41.41 ± 10.66 | 13.33 ± 2.16 | 0.24 ± 0.05 | |
| APOC3 rs4520 | C/C | 16 | 682.54 ± 217.81 | 4610.74 ± 1129.47 | 2.47 ± 0.75 | 17.57 ± 4.14 | 119.57 ± 22.68 | 47.08 ± 10.47 * | 14.92 ± 2.77 | 0.23 ± 0.05 |
| C/T | 9 | 598.48 ± 190.61 | 3957.67 ± 793.57 | 2 ± 0.5 | 20.63 ± 6.17 | 136.9 ± 27.08 | 36.08 ± 5.97 * | 13.58 ± 3.39 | 0.26 ± 0.05 | |
| APOC3 rs5128 | C/C | 20 | 670.73 ± 209.34 | 4476.06 ± 1081.81 | 2.33 ± 0.74 | 18.31 ± 4.87 | 122.79 ± 25.84 | 45.49 ± 10.11 * | 14.93 ± 3.05 | 0.24 ± 0.05 |
| C/G | 5 | 578.48 ± 209.97 | 3973.95 ± 919.99 | 2.2 ± 0.57 | 20.09 ± 6.17 | 137.88 ± 20.7 | 33.63 ± 5.45 * | 12.48 ± 2 | 0.26 ± 0.06 | |
| COMT rs4680 | G/G | 12 | 558.64 ± 193.87 * | 3786.77 ± 761.8 * | 2.21 ± 0.53 | 18.13 ± 4.47 | 124.78 ± 20.6 | 37.03 ± 7.25 * | 14.34 ± 2.81 | 0.27 ± 0.05 * |
| G/A | 13 | 738.71 ± 188.69 * | 4919.21 ± 1010.12 * | 2.38 ± 0.84 | 19.17 ± 5.7 | 126.76 ± 29.75 | 48.74 ± 9.96 * | 14.53 ± 3.29 | 0.21 ± 0.04 * | |
| CYP2A6 rs28399433 | A/A | 19 | 615.71 ± 180.08 | 4077.07 ± 865.3 *,#2 | 2.26 ± 0.76 | 18.77 ± 5.03 | 124.82 ± 25.52 | 41.61 ± 9.25 | 14.93 ± 3.08 | 0.26 ± 0.05 *,#2 |
| A/C | 4 | 872.46 ± 243.77 | 5557.56 ± 1128 * | 2.38 ± 0.48 | 19.6 ± 6.71 | 123.7 ± 32.52 | 52.05 ± 12.33 | 13.54 ± 2.49 | 0.19 ± 0.03 * | |
| C/C | 2 | 559.35 ± 210.72 | 4848.16 ± 1267.87 * | 2.5 ± 0.71 | 15.86 ± 2.27 | 139.46 ± 3.08 | 39.61 ± 15.6 | 11.53 ± 1.56 | 0.22 ± 0.05 * | |
| CYP2C9 | NM | 21 | 631.89 ± 214.21 | 4364.55 ± 1119.04 | 2.38 ± 0.73 | 17.52 ± 4.32 * | 122.36 ± 25.34 | 43.68 ± 11.23 | 14.62 ± 2.99 | 0.24 ± 0.06 |
| IM | 4 | 759.31 ± 153.58 | 4433.81 ± 723.12 | 1.88 ± 0.25 | 24.7 ± 4.79 * | 143.94 ± 17.41 | 40.17 ± 4.54 | 13.46 ± 3.34 | 0.23 ± 0.03 | |
| CYP2D6 | UM + NM | 21 | 591.81 ± 152.02 * | 4093.18 ± 820.11 * | 2.31 ± 0.76 | 18.22 ± 5.32 | 125.34 ± 26.43 | 40.62 ± 8.90 * | 14.56 ± 3.26 | 0.25 ± 0.05 * |
| IM + PM | 4 | 969.71 ± 183.48 * | 5858.51 ± 950.00 * | 2.25 ± 0.29 | 21.03 ± 2.72 | 128.28 ± 25.26 | 56.22 ± 8.61 * | 13.80 ± 0.92 | 0.18 ± 0.03 * | |
| DRD3 rs6280 | T/T | 6 | 710.97 ± 154.29 | 4522.5 ± 782.39 | 1.67 ± 0.26 *,#3 | 23.09 ± 5.83 | 144.42 ± 18.5 *,#4 | 40.8 ± 10 | 13.03 ± 2.34 | 0.23 ± 0.04 |
| T/C | 10 | 651.34 ± 248.12 | 4135.9 ± 1011.25 | 2.35 ± 0.66 * | 17.2 ± 4.06 | 111.81 ± 17.16 * | 43.97 ± 8.43 | 15.57 ± 2.15 | 0.26 ± 0.06 | |
| C/C | 9 | 614.19 ± 204.99 | 4544.1 ± 1294.52 | 2.67 ± 0.7 * | 17.36 ± 4.23 | 128.96 ± 29.21 * | 43.72 ± 13.43 | 14.12 ± 3.91 | 0.24 ± 0.06 | |
| HTR2A rs6313 | C/C | 12 | 665.26 ± 226.85 | 4376.74 ± 1232.73 | 2.02 ± 0.41 *,#5 | 20.1 ± 5.34 *,#5 | 132.9 ± 26.24 *,#5 | 42.17 ± 10.6 | 14.17 ± 2.59 | 0.25 ± 0.06 |
| C/T | 9 | 683.34 ± 220 | 4544.29 ± 1003.05 | 2.22 ± 0.74∗ | 19.22 ± 4.16 * | 128.59 ± 15.79 * | 44.2 ± 11.01 | 14.09 ± 2.34 | 0.23 ± 0.05 | |
| T/T | 4 | 543.42 ± 102.69 | 3992.87 ± 584.04 | 3.31 ± 0.38 *,#6 | 13.15 ± 2.24 * | 98.3 ± 26.12 * | 43.52 ± 11.52 | 16.02 ± 5.4 | 0.26 ± 0.04 | |
| HTR2C rs518147 | C/C | 11 | 522.27 ± 145.24 *,#7 | 4047.75 ± 797.42 | 2.32 ± 0.55 | 16.57 ± 4.62 | 127.92 ± 25.94 | 38.24 ± 10.4 | 13.63 ± 2.5 | 0.26 ± 0.05 |
| C/G | 6 | 779.75 ± 244.12 * | 4566.77 ± 1444.99 | 2.21 ± 0.84 | 20.13 ± 6.45 | 116.95 ± 29.85 | 46.79 ± 11.48 | 15.23 ± 3.03 | 0.24 ± 0.07 | |
| G/G | 8 | 735.43 ± 167.05 *,#8 | 4683.12 ± 1046.98 | 2.34 ± 0.86 | 20.46 ± 3.92 | 129.55 ± 22.35 | 47.07 ± 7.72 | 14.96 ± 3.69 | 0.23 ± 0.05 | |
| Total | 25 | 652.28 ± 208.47 | 4375.64 ± 1053.37 | 2.3 ± 0.7 | 18.67 ± 5.07 | 125.81 ± 25.26 | 43.12 ± 10.46 | 14.44 ± 3.01 | 0.24 ± 0.05 |
| Drug | Pharmacokinetic Parameter | No AEs (n = 10) | AEs (n = 15) | Total (n = 25) | p-Value |
|---|---|---|---|---|---|
| Donepezil | AUC (ng∗h/mL) | 583.12 ± 174.53 | 698.38 ± 221.86 | 652.28 ± 208.47 | 0.175 |
| AUC/DW (ng∗h∗mg/mL∗kg) | 4308.93 ± 992.26 | 4420.11 ± 1124.23 | 4375.64 ± 1053.37 | 0.822 | |
| Tmax (h) | 2.38 ± 0.78 | 2.25 ± 0.66 | 2.30 ± 0.70 | 0.747 | |
| Cmax (pg/mL) | 16.26 ± 4.60 | 20.28 ± 4.84 | 18.67 ± 5.07 | 0.041 * | |
| Cmax/DW (ng∗mg/mL∗kg) | 119.32 ± 20.2 | 130.14 ± 27.95 | 125.81 ± 25.26 | 0.382 | |
| Vd/F (L/kg) | 14.47 ± 3.16 | 14.42 ± 3.01 | 14.44 ± 3.01 | 0.988 | |
| Cl/F (L/h∗kg) | 0.25 ± 0.06 | 0.24 ± 0.05 | 0.24 ± 0.05 | 0.824 | |
| T½ (h) | 42.67 ± 10.72 | 43.42 ± 10.65 | 43.12 ± 10.46 | 0.837 | |
| Memantine | AUC (ng∗h/mL) | 901.07 ± 216.56 | 1106.09 ± 266.92 | 1024.08 ± 263.92 | 0.049 |
| AUC/DW (ng∗h∗mg/mL∗kg) | 6683.05 ± 1092.15 | 7043.73 ± 1368.14 | 6899.46 ± 1253.68 | 0.504 | |
| Tmax (h) | 5.68 ± 1.93 | 5.4 ± 2.38 | 5.51 ± 2.18 | 0.581 | |
| Cmax (ng/mL) | 13.46 ± 2.75 | 17.07 ± 3.35 | 15.63 ± 3.55 | 0.007 * | |
| Cmax/DW (ng∗mg/mL∗kg) | 99.39 ± 8.14 | 108.91 ± 13.89 | 105.1 ± 12.65 | 0.084 | |
| Vd/F (L/kg) | 9.84 ± 0.92 | 9.37 ± 1.12 | 9.56 ± 1.05 | 0.254 | |
| Cl/F (L/h∗kg) | 0.15 ± 0.03 | 0.15 ± 0.03 | 0.15 ± 0.03 | 0.501 | |
| T½ (h) | 45.69 ± 9.44 | 45.56 ± 9 | 45.61 ± 8.98 | 0.980 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ovejero-Benito, M.C.; Ochoa, D.; Enrique-Benedito, T.; del Peso-Casado, M.; Zubiaur, P.; Navares, M.; Román, M.; Abad-Santos, F. Pharmacogenetics of Donepezil and Memantine in Healthy Subjects. J. Pers. Med. 2022, 12, 788. https://doi.org/10.3390/jpm12050788
Ovejero-Benito MC, Ochoa D, Enrique-Benedito T, del Peso-Casado M, Zubiaur P, Navares M, Román M, Abad-Santos F. Pharmacogenetics of Donepezil and Memantine in Healthy Subjects. Journal of Personalized Medicine. 2022; 12(5):788. https://doi.org/10.3390/jpm12050788
Chicago/Turabian StyleOvejero-Benito, María C., Dolores Ochoa, Teresa Enrique-Benedito, Miriam del Peso-Casado, Pablo Zubiaur, Marcos Navares, Manuel Román, and Francisco Abad-Santos. 2022. "Pharmacogenetics of Donepezil and Memantine in Healthy Subjects" Journal of Personalized Medicine 12, no. 5: 788. https://doi.org/10.3390/jpm12050788