Liquid Biopsy Biomarkers in Urine: A Route towards Molecular Diagnosis and Personalized Medicine of Bladder Cancer
Abstract
:1. Introduction
2. Methods
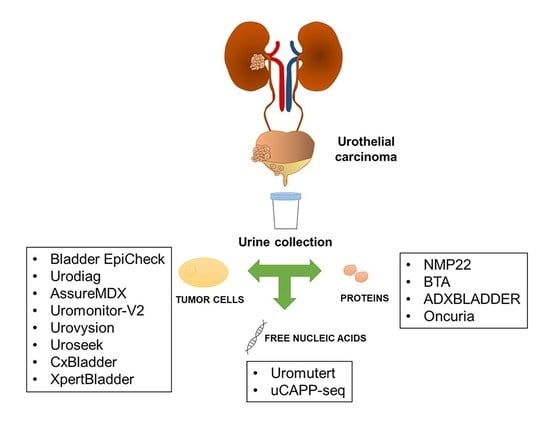
3. Biomarkers Used in Urinary Biopsy
4. Molecular Detection and Surveillance
5. DNA Methylation
5.1. Bladder EpiCheck
5.2. Urodiag
5.3. AssureMDX
6. DNA Mutations
6.1. UroMuTERT
6.2. Uromonitor
6.3. Urovysion
6.4. Uroseek
6.5. uCAPP-Seq
7. Protein-Based Assays
NMP22 and Basement Membrane-Derived Antigen (BTA)
8. Adxbladder
Oncuria
9. Non-Coding RNA
10. mRNA Signatures
10.1. Cxbladder Detect and Monitor
10.2. Xpert Bladder Cancer Detect and Monitor
11. Predict Targeted Therapy Efficacy
12. Conclusions and Future Perspective
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
References
- Murtaza, M.; Dawson, S.J.; Tsui, D.W.; Gale, D.; Forshew, T.; Piskorz, A.M.; Parkinson, C.; Chin, S.F.; Kingsbury, Z.; Wong, A.S.; et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013, 497, 108–112. [Google Scholar] [CrossRef]
- Chen, L.; Bode, A.M.; Dong, Z. Circulating tumor cells: Moving biological insights into detection. Theranostics 2017, 7, 2606–2619. [Google Scholar] [CrossRef]
- Satyal, U.; Srivastava, A.; Abbosh, P.H. Urine biopsy-liquid gold for molecular detection and surveillance of bladder cancer. Front. Oncol. 2019, 9, 1266. [Google Scholar] [CrossRef] [Green Version]
- Normanno, N.; Cervantes, A.; Ciardiello, F.; De Luca, A.; Pinto, C. The liquid biopsy in the management of colorectal cancer patients: Current applications and future scenarios. Cancer Treat. Rev. 2018, 70, 1–8. [Google Scholar] [CrossRef]
- Boonham, N.; Kreuze, J.; Winter, S.; van der Vlugt, R.; Bergervoet, J.; Tomlinson, J.; Mumford, R. Methods in virus diagnostics: From ELISA to next generation sequencing. Virus Res. 2014, 186, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Mischak, H.; Delles, C.; Vlahou, A.; Vanholder, R. Proteomic biomarkers in kidney disease: Issues in development and implementation. Nat. Rev. Nephrol. 2015, 11, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Rzhevskiy, A.S.; Razavi Bazaz, S.; Ding, L.; Kapitannikova, A.; Sayyadi, N.; Campbell, D.; Walsh, B.; Gillatt, D.; Ebrahimi Warkiani, M.; Zvyagin, A.V. Rapid and label-free isolation of tumour cells from the urine of patients with localised prostate cancer using inertial microfluidics. Cancers 2019, 12, 81. [Google Scholar] [CrossRef] [Green Version]
- Laguna, T.A.; Wagner, B.D.; Starcher, B.; Luckey Tarro, H.K.; Mann, S.A.; Sagel, S.D.; Accurso, F.J. Urinary desmosine: A biomarker of structural lung injury during CF pulmonary exacerbation. Pediatr. Pulmonol. 2012, 47, 856–863. [Google Scholar] [CrossRef] [Green Version]
- Ongay, S.; Sikma, M.; Horvatovich, P.; Hermans, J.; Miller, B.E.; Ten Hacken, N.H.T.; Bischoff, R. Free urinary desmosine and isodesmosine as COPD biomarkers: The relevance of confounding factors. Chronic Obstr. Pulm. Dis. 2016, 3, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Togneri, F.S.; Ward, D.G.; Foster, J.M.; Devall, A.J.; Wojtowicz, P.; Alyas, S.; Vasques, F.R.; Oumie, A.; James, N.D.; Cheng, K.K.; et al. Genomic complexity of urothelial bladder cancer revealed in urinary cfDNA. Eur. J. Hum. Genet. 2016, 24, 1167–1174. [Google Scholar] [CrossRef] [Green Version]
- Choi, H.S.; Liu, W.; Misra, P.; Tanaka, E.; Zimmer, J.P.; Itty Ipe, B.; Bawendi, M.G.; Frangioni, J.V. Renal clearance of quantum dots. Nat. Biotechnol. 2007, 25, 1165–1170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef]
- Lodewijk, I.; Duenas, M.; Rubio, C.; Munera-Maravilla, E.; Segovia, C.; Bernardini, A.; Teijeira, A.; Paramio, J.M.; Suarez-Cabrera, C. Liquid biopsy biomarkers in bladder cancer: A current need for patient diagnosis and monitoring. Int. J. Mol. Sci. 2018, 19, 2514. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.J.; Reich, C.F., 3rd; Pisetsky, D.S. The role of macrophages in the in vitro generation of extracellular DNA from apoptotic and necrotic cells. Immunology 2005, 115, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Bryzgunova, O.E.; Skvortsova, T.E.; Kolesnikova, E.V.; Starikov, A.V.; Rykova, E.Y.; Vlassov, V.V.; Laktionov, P.P. Isolation and comparative study of cell-free nucleic acids from human urine. Ann. N. Y. Acad. Sci. 2006, 1075, 334–340. [Google Scholar] [CrossRef]
- Phallen, J.; Sausen, M.; Adleff, V.; Leal, A.; Hruban, C.; White, J.; Anagnostou, V.; Fiksel, J.; Cristiano, S.; Papp, E.; et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirotsu, Y.; Yokoyama, H.; Amemiya, K.; Hagimoto, T.; Daimon, H.; Hosaka, K.; Oyama, T.; Mochizuki, H.; Omata, M. Genomic profile of urine has high diagnostic sensitivity compared to cytology in non-invasive urothelial bladder cancer. Cancer Sci. 2019, 110, 3235–3243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, C.P.; Finney, H. Developments in the assessment of glomerular filtration rate. Clin. Chim. Acta 2000, 297, 55–66. [Google Scholar] [CrossRef]
- D’Costa, J.J.; Goldsmith, J.C.; Wilson, J.S.; Bryan, R.T.; Ward, D.G. A systematic review of the diagnostic and prognostic value of urinary protein biomarkers in urothelial bladder cancer. Bladder Cancer 2016, 2, 301–317. [Google Scholar] [CrossRef] [Green Version]
- Krochmal, M.; van Kessel, K.E.M.; Zwarthoff, E.C.; Belczacka, I.; Pejchinovski, M.; Vlahou, A.; Mischak, H.; Frantzi, M. Urinary peptide panel for prognostic assessment of bladder cancer relapse. Sci. Rep. 2019, 9, 7635. [Google Scholar] [CrossRef] [Green Version]
- Frantzi, M.; van Kessel, K.E.; Zwarthoff, E.C.; Marquez, M.; Rava, M.; Malats, N.; Merseburger, A.S.; Katafigiotis, I.; Stravodimos, K.; Mullen, W.; et al. Development and validation of urine-based peptide biomarker panels for detecting bladder cancer in a multi-center study. Clin. Cancer Res. 2016, 22, 4077–4086. [Google Scholar] [CrossRef] [Green Version]
- Hussein, A.A.; Elsayed, A.S.; Durrani, M.; Jing, Z.; Iqbal, U.; Gomez, E.C.; Singh, P.K.; Liu, S.; Smith, G.; Tang, L.; et al. Investigating the association between the urinary microbiome and bladder cancer: An exploratory study. Urol. Oncol. 2021. [Google Scholar] [CrossRef]
- Wu, P.; Zhang, G.; Zhao, J.; Chen, J.; Chen, Y.; Huang, W.; Zhong, J.; Zeng, J. Profiling the urinary microbiota in male patients with bladder cancer in China. Front. Cell Infect. Microbiol. 2018, 8, 167. [Google Scholar] [CrossRef] [PubMed]
- Roupret, M.; Babjuk, M.; Burger, M.; Capoun, O.; Cohen, D.; Comperat, E.M.; Cowan, N.C.; Dominguez-Escrig, J.L.; Gontero, P.; Hugh Mostafid, A.; et al. European Association of Urology Guidelines on upper urinary tract urothelial carcinoma: 2020 update. Eur. Urol. 2021, 79, 62–79. [Google Scholar] [CrossRef] [PubMed]
- Burger, M.; Catto, J.W.; Dalbagni, G.; Grossman, H.B.; Herr, H.; Karakiewicz, P.; Kassouf, W.; Kiemeney, L.A.; La Vecchia, C.; Shariat, S.; et al. Epidemiology and risk factors of urothelial bladder cancer. Eur. Urol. 2013, 63, 234–241. [Google Scholar] [CrossRef]
- Van Kessel, K.E.; Beukers, W.; Lurkin, I.; Ziel-van der Made, A.; van der Keur, K.A.; Boormans, J.L.; Dyrskjot, L.; Marquez, M.; Orntoft, T.F.; Real, F.X.; et al. Validation of a DNA methylation-mutation urine assay to select patients with hematuria for cystoscopy. J. Urol. 2017, 197, 590–595. [Google Scholar] [CrossRef]
- Beukers, W.; Kandimalla, R.; Masius, R.G.; Vermeij, M.; Kranse, R.; van Leenders, G.J.; Zwarthoff, E.C. Stratification based on methylation of TBX2 and TBX3 into three molecular grades predicts progression in patients with pTa-bladder cancer. Mod. Pathol. 2015, 28, 515–522. [Google Scholar] [CrossRef] [Green Version]
- Mancini, M.; Righetto, M.; Zumerle, S.; Montopoli, M.; Zattoni, F. The Bladder EpiCheck Test as a non-invasive tool based on the identification of DNA methylation in bladder cancer cells in the urine: A review of published evidence. Int. J. Mol. Sci. 2020, 21, 6542. [Google Scholar] [CrossRef] [PubMed]
- Trenti, E.; D’Elia, C.; Mian, C.; Schwienbacher, C.; Hanspeter, E.; Pycha, A.; Kafka, M.; Degener, S.; Danuser, H.; Roth, S.; et al. Diagnostic predictive value of the Bladder EpiCheck test in the follow-up of patients with non-muscle-invasive bladder cancer. Cancer Cytopathol. 2019, 127, 465–469. [Google Scholar] [CrossRef]
- D’Andrea, D.; Soria, F.; Zehetmayer, S.; Gust, K.M.; Korn, S.; Witjes, J.A.; Shariat, S.F. Diagnostic accuracy, clinical utility and influence on decision-making of a methylation urine biomarker test in the surveillance of non-muscle-invasive bladder cancer. BJU Int. 2019, 123, 959–967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trenti, E.; Pycha, S.; Mian, C.; Schwienbacher, C.; Hanspeter, E.; Kafka, M.; Spedicato, G.A.; Vjaters, E.; Degener, S.; Pycha, A.; et al. Comparison of 2 new real-time polymerase chain reaction-based urinary markers in the follow-up of patients with non-muscle-invasive bladder cancer. Cancer Cytopathol. 2020, 128, 341–347. [Google Scholar] [CrossRef]
- Witjes, J.A.; Morote, J.; Cornel, E.B.; Gakis, G.; van Valenberg, F.J.P.; Lozano, F.; Sternberg, I.A.; Willemsen, E.; Hegemann, M.L.; Paitan, Y.; et al. Performance of the bladder EpiCheck methylation test for patients under surveillance for non-muscle-invasive bladder cancer: Results of a multicenter, prospective, blinded clinical trial. Eur. Urol. Oncol. 2018, 1, 307–313. [Google Scholar] [CrossRef]
- Brait, M.; Begum, S.; Carvalho, A.L.; Dasgupta, S.; Vettore, A.L.; Czerniak, B.; Caballero, O.L.; Westra, W.H.; Sidransky, D.; Hoque, M.O. Aberrant promoter methylation of multiple genes during pathogenesis of bladder cancer. Cancer Epidemiol. Biomark. Prev. 2008, 17, 2786–2794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roperch, J.P.; Hennion, C. A novel ultra-sensitive method for the detection of FGFR3 mutations in urine of bladder cancer patients—Design of the Urodiag(R) PCR kit for surveillance of patients with non-muscle-invasive bladder cancer (NMIBC). BMC Med. Genet. 2020, 21, 112. [Google Scholar] [CrossRef] [PubMed]
- Roperch, J.P.; Grandchamp, B.; Desgrandchamps, F.; Mongiat-Artus, P.; Ravery, V.; Ouzaid, I.; Roupret, M.; Phe, V.; Ciofu, C.; Tubach, F.; et al. Promoter hypermethylation of HS3ST2, SEPTIN9 and SLIT2 combined with FGFR3 mutations as a sensitive/specific urinary assay for diagnosis and surveillance in patients with low or high-risk non-muscle-invasive bladder cancer. BMC Cancer 2016, 16, 704. [Google Scholar] [CrossRef]
- Van Kessel, K.E.; Van Neste, L.; Lurkin, I.; Zwarthoff, E.C.; Van Criekinge, W. Evaluation of an epigenetic profile for the detection of bladder cancer in patients with hematuria. J. Urol. 2016, 195, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Beukers, W.; van der Keur, K.A.; Kandimalla, R.; Vergouwe, Y.; Steyerberg, E.W.; Boormans, J.L.; Jensen, J.B.; Lorente, J.A.; Real, F.X.; Segersten, U.; et al. FGFR3, TERT and OTX1 as a urinary biomarker combination for surveillance of patients with bladder cancer in a large prospective multicenter study. J. Urol. 2017, 197, 1410–1418. [Google Scholar] [CrossRef] [PubMed]
- Hosen, I.; Rachakonda, P.S.; Heidenreich, B.; de Verdier, P.J.; Ryk, C.; Steineck, G.; Hemminki, K.; Kumar, R. Mutations in TERT promoter and FGFR3 and telomere length in bladder cancer. Int. J. Cancer 2015, 137, 1621–1629. [Google Scholar] [CrossRef] [PubMed]
- Kinde, I.; Munari, E.; Faraj, S.F.; Hruban, R.H.; Schoenberg, M.; Bivalacqua, T.; Allaf, M.; Springer, S.; Wang, Y.; Diaz, L.A., Jr.; et al. TERT promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in urine. Cancer Res. 2013, 73, 7162–7167. [Google Scholar] [CrossRef] [Green Version]
- Avogbe, P.H.; Manel, A.; Vian, E.; Durand, G.; Forey, N.; Voegele, C.; Zvereva, M.; Hosen, M.I.; Meziani, S.; De Tilly, B.; et al. Urinary TERT promoter mutations as non-invasive biomarkers for the comprehensive detection of urothelial cancer. EBioMedicine 2019, 44, 431–438. [Google Scholar] [CrossRef] [Green Version]
- Hosen, M.I.; Sheikh, M.; Zvereva, M.; Scelo, G.; Forey, N.; Durand, G.; Voegele, C.; Poustchi, H.; Khoshnia, M.; Roshandel, G.; et al. Urinary TERT promoter mutations are detectable up to 10 years prior to clinical diagnosis of bladder cancer: Evidence from the Golestan Cohort Study. EBioMedicine 2020, 53, 102643. [Google Scholar] [CrossRef]
- Hosen, M.I.; Forey, N.; Durand, G.; Voegele, C.; Bilici, S.; Avogbe, P.H.; Delhomme, T.M.; Foll, M.; Manel, A.; Vian, E.; et al. Development of sensitive droplet digital PCR assays for detecting urinary TERT promoter mutations as non-invasive biomarkers for detection of urothelial cancer. Cancers 2020, 12, 3541. [Google Scholar] [CrossRef]
- Hernandez, S.; Lopez-Knowles, E.; Lloreta, J.; Kogevinas, M.; Amoros, A.; Tardon, A.; Carrato, A.; Serra, C.; Malats, N.; Real, F.X. Prospective study of FGFR3 mutations as a prognostic factor in nonmuscle invasive urothelial bladder carcinomas. J. Clin. Oncol. 2006, 24, 3664–3671. [Google Scholar] [CrossRef]
- Sieverink, C.A.; Batista, R.P.M.; Prazeres, H.J.M.; Vinagre, J.; Sampaio, C.; Leao, R.R.; Maximo, V.; Witjes, J.A.; Soares, P. Clinical validation of a urine test (uromonitor-V2((R))) for the surveillance of non-muscle-invasive bladder cancer patients. Diagnostics 2020, 10, 745. [Google Scholar] [CrossRef] [PubMed]
- Hajdinjak, T. UroVysion FISH test for detecting urothelial cancers: Meta-analysis of diagnostic accuracy and comparison with urinary cytology testing. Urol. Oncol. 2008, 26, 646–651. [Google Scholar] [CrossRef]
- Dimashkieh, H.; Wolff, D.J.; Smith, T.M.; Houser, P.M.; Nietert, P.J.; Yang, J. Evaluation of urovysion and cytology for bladder cancer detection: A study of 1835 paired urine samples with clinical and histologic correlation. Cancer Cytopathol. 2013, 121, 591–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halling, K.C.; Kipp, B.R. Bladder cancer detection using FISH (UroVysion assay). Adv. Anat. Pathol. 2008, 15, 279–286. [Google Scholar] [CrossRef]
- Whitson, J.; Berry, A.; Carroll, P.; Konety, B. A multicolour fluorescence in situ hybridization test predicts recurrence in patients with high-risk superficial bladder tumours undergoing intravesical therapy. BJU Int. 2009, 104, 336–339. [Google Scholar] [CrossRef]
- Savic, S.; Zlobec, I.; Thalmann, G.N.; Engeler, D.; Schmauss, M.; Lehmann, K.; Mattarelli, G.; Eichenberger, T.; Dalquen, P.; Spieler, P.; et al. The prognostic value of cytology and fluorescence in situ hybridization in the follow-up of nonmuscle-invasive bladder cancer after intravesical Bacillus Calmette-Guerin therapy. Int. J. Cancer 2009, 124, 2899–2904. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.; Rosser, C.J. UroSEEK gene panel for bladder cancer surveillance. Transl. Androl. Urol. 2019, 8, S546–S549. [Google Scholar] [CrossRef]
- Eich, M.L.; Rodriguez Pena, M.D.C.; Springer, S.U.; Taheri, D.; Tregnago, A.C.; Salles, D.C.; Bezerra, S.M.; Cunha, I.W.; Fujita, K.; Ertoy, D.; et al. Incidence and distribution of UroSEEK gene panel in a multi-institutional cohort of bladder urothelial carcinoma. Mod. Pathol. 2019, 32, 1544–1550. [Google Scholar] [CrossRef]
- Dudley, J.C.; Schroers-Martin, J.; Lazzareschi, D.V.; Shi, W.Y.; Chen, S.B.; Esfahani, M.S.; Trivedi, D.; Chabon, J.J.; Chaudhuri, A.A.; Stehr, H.; et al. Detection and surveillance of bladder cancer using urine tumor DNA. Cancer Discov 2019, 9, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Carpinito, G.A.; Stadler, W.M.; Briggman, J.V.; Chodak, G.W.; Church, P.A.; Lamm, D.L.; Lange, P.H.; Messing, E.M.; Pasciak, R.M.; Reservitz, G.B.; et al. Urinary nuclear matrix protein as a marker for transitional cell carcinoma of the urinary tract. J. Urol. 1996, 156, 1280–1285. [Google Scholar] [CrossRef]
- Mowatt, G.; Zhu, S.; Kilonzo, M.; Boachie, C.; Fraser, C.; Griffiths, T.R.; N’Dow, J.; Nabi, G.; Cook, J.; Vale, L. Systematic review of the clinical effectiveness and cost-effectiveness of photodynamic diagnosis and urine biomarkers (FISH, ImmunoCyt, NMP22) and cytology for the detection and follow-up of bladder cancer. Health Technol. Assess. 2010, 14, 3–4. [Google Scholar] [CrossRef] [Green Version]
- Soria, F.; Droller, M.J.; Lotan, Y.; Gontero, P.; D’Andrea, D.; Gust, K.M.; Roupret, M.; Babjuk, M.; Palou, J.; Shariat, S.F. An up-to-date catalog of available urinary biomarkers for the surveillance of non-muscle invasive bladder cancer. World J. Urol. 2018, 36, 1981–1995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyake, M.; Goodison, S.; Rizwani, W.; Ross, S.; Bart Grossman, H.; Rosser, C.J. Urinary BTA: Indicator of bladder cancer or of hematuria. World J. Urol. 2012, 30, 869–873. [Google Scholar] [CrossRef] [Green Version]
- Lokeshwar, V.B.; Soloway, M.S. Current bladder tumor tests: Does their projected utility fulfill clinical necessity? J. Urol. 2001, 165, 1067–1077. [Google Scholar] [CrossRef]
- Kinders, R.; Jones, T.; Root, R.; Bruce, C.; Murchison, H.; Corey, M.; Williams, L.; Enfield, D.; Hass, G.M. Complement factor H or a related protein is a marker for transitional cell cancer of the bladder. Clin. Cancer Res. 1998, 4, 2511–2520. [Google Scholar]
- Sarosdy, M.F.; deVere White, R.W.; Soloway, M.S.; Sheinfeld, J.; Hudson, M.A.; Schellhammer, P.F.; Jarowenko, M.V.; Adams, G.; Blumenstein, B.A. Results of a multicenter trial using the BTA test to monitor for and diagnose recurrent bladder cancer. J. Urol. 1995, 154, 379–383. [Google Scholar] [CrossRef]
- Tye, B.K. MCM proteins in DNA replication. Ann. Rev. Biochem. 1999, 68, 649–686. [Google Scholar] [CrossRef]
- Stoeber, K.; Tlsty, T.D.; Happerfield, L.; Thomas, G.A.; Romanov, S.; Bobrow, L.; Williams, E.D.; Williams, G.H. DNA replication licensing and human cell proliferation. J. Cell Sci. 2001, 114, 2027–2041. [Google Scholar]
- Stoeber, K.; Halsall, I.; Freeman, A.; Swinn, R.; Doble, A.; Morris, L.; Coleman, N.; Bullock, N.; Laskey, R.A.; Hales, C.N.; et al. Immunoassay for urothelial cancers that detects DNA replication protein Mcm5 in urine. Lancet 1999, 354, 1524–1525. [Google Scholar] [CrossRef]
- Stoeber, K.; Swinn, R.; Prevost, A.T.; de Clive-Lowe, P.; Halsall, I.; Dilworth, S.M.; Marr, J.; Turner, W.H.; Bullock, N.; Doble, A.; et al. Diagnosis of genito-urinary tract cancer by detection of minichromosome maintenance 5 protein in urine sediments. J. Natl. Cancer Inst. 2002, 94, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.D.; Dudderidge, T.J.; Wollenschlaeger, A.; Okoturo, O.; Burling, K.; Tulloch, F.; Halsall, I.; Prevost, T.; Prevost, A.T.; Vasconcelos, J.C.; et al. Bladder cancer diagnosis and identification of clinically significant disease by combined urinary detection of Mcm5 and nuclear matrix protein 22. PLoS ONE 2012, 7, e40305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudderidge, T.; Stockley, J.; Nabi, G.; Mom, J.; Umez-Eronini, N.; Hrouda, D.; Cresswell, J.; McCracken, S.R.C. A Novel, non-invasive test enabling bladder cancer detection in urine sediment of patients presenting with haematuria-A prospective multicentre performance evaluation of ADXBLADDER. Eur. Urol. Oncol. 2020, 3, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Anastasi, E.; Maggi, M.; Tartaglione, S.; Angeloni, A.; Gennarini, G.; Leoncini, P.P.; Sperduti, I.; Di Lascio, G.; De Stefano, V.; Di Pierro, G.B.; et al. Predictive value of MCM5 (ADXBLADDER) analysis in urine of men evaluated for the initial diagnosis of bladder cancer: A comparative prospective study. Diagn. Cytopathol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Roupret, M.; Gontero, P.; McCracken, S.R.C.; Dudderidge, T.; Stockley, J.; Kennedy, A.; Rodriguez, O.; Sieverink, C.; Vanie, F.; Allasia, M.; et al. Diagnostic accuracy of MCM5 for the detection of recurrence in nonmuscle invasive bladder cancer followup: A blinded, prospective cohort, multicenter European Study. J. Urol. 2020, 204, 685–690. [Google Scholar] [CrossRef]
- Furuya, H.; Tabula, L.; Lee, R.; Kralovec, P.; Ramsden, M.; Wong, R.; Rosser, C.J. Analytical validation of ONCURIA a multiplex bead-based immunoassay for the non-invasive bladder cancer detection. Pract. Lab. Med. 2020, 22, e00189. [Google Scholar] [CrossRef]
- Yazarlou, F.; Modarressi, M.H.; Mowla, S.J.; Oskooei, V.K.; Motevaseli, E.; Tooli, L.F.; Nekoohesh, L.; Eghbali, M.; Ghafouri-Fard, S.; Afsharpad, M. Urinary exosomal expression of long non-coding RNAs as diagnostic marker in bladder cancer. Cancer Manag. Res. 2018, 10, 6357–6365. [Google Scholar] [CrossRef] [Green Version]
- Parizi, P.K.; Yarahmadi, F.; Tabar, H.M.; Hosseini, Z.; Sarli, A.; Kia, N.; Tafazoli, A.; Esmaeili, S.A. MicroRNAs and target molecules in bladder cancer. Med. Oncol. 2020, 37, 118. [Google Scholar] [CrossRef]
- Hanke, M.; Hoefig, K.; Merz, H.; Feller, A.C.; Kausch, I.; Jocham, D.; Warnecke, J.M.; Sczakiel, G. A robust methodology to study urine microRNA as tumor marker: MicroRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol. Oncol. 2010, 28, 655–661. [Google Scholar] [CrossRef]
- Flippot, R.; Beinse, G.; Boileve, A.; Vibert, J.; Malouf, G.G. Long non-coding RNAs in genitourinary malignancies: A whole new world. Nat. Rev. Urol. 2019, 16, 484–504. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Singh, P.K.; Rath, S.K.; Dalela, D.; Goel, M.M.; Bhatt, M.L. Appraisal of diagnostic ability of UCA1 as a biomarker of carcinoma of the urinary bladder. Tumour Biol. 2014, 35, 11435–11442. [Google Scholar] [CrossRef]
- Wang, X.S.; Zhang, Z.; Wang, H.C.; Cai, J.L.; Xu, Q.W.; Li, M.Q.; Chen, Y.C.; Qian, X.P.; Lu, T.J.; Yu, L.Z.; et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin. Cancer Res. 2006, 12, 4851–4858. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wang, X.; Zhang, D.; Yu, Y.; Cai, L.; Zhang, C. Long non-coding RNA urothelial carcinoma-associated 1 as a tumor biomarker for the diagnosis of urinary bladder cancer. Tumour Biol. 2017, 39, 1010428317709990. [Google Scholar] [CrossRef] [Green Version]
- O’Sullivan, P.; Sharples, K.; Dalphin, M.; Davidson, P.; Gilling, P.; Cambridge, L.; Harvey, J.; Toro, T.; Giles, N.; Luxmanan, C.; et al. A multigene urine test for the detection and stratification of bladder cancer in patients presenting with hematuria. J. Urol. 2012, 188, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Kavalieris, L.; O’Sullivan, P.; Frampton, C.; Guilford, P.; Darling, D.; Jacobson, E.; Suttie, J.; Raman, J.D.; Shariat, S.F.; Lotan, Y. Performance characteristics of a multigene urine biomarker test for monitoring for recurrent urothelial carcinoma in a multicenter study. J. Urol. 2017, 197, 1419–1426. [Google Scholar] [CrossRef] [PubMed]
- Lotan, Y.; O’Sullivan, P.; Raman, J.D.; Shariat, S.F.; Kavalieris, L.; Frampton, C.; Guilford, P.; Luxmanan, C.; Suttie, J.; Crist, H.; et al. Clinical comparison of noninvasive urine tests for ruling out recurrent urothelial carcinoma. Urol. Oncol. 2017, 35, e515–e531. [Google Scholar] [CrossRef]
- Koya, M.; Osborne, S.; Chemasle, C.; Porten, S.; Schuckman, A.; Kennedy-Smith, A. An evaluation of the real world use and clinical utility of the Cxbladder Monitor assay in the follow-up of patients previously treated for bladder cancer. BMC Urol. 2020, 20, 12. [Google Scholar] [CrossRef] [PubMed]
- Wallace, E.; Higuchi, R.; Satya, M.; McCann, L.; Sin, M.L.Y.; Bridge, J.A.; Wei, H.; Zhang, J.; Wong, E.; Hiar, A.; et al. Development of a 90-minute integrated noninvasive urinary assay for bladder cancer detection. J. Urol. 2018, 199, 655–662. [Google Scholar] [CrossRef]
- Valenberg, F.; Hiar, A.M.; Wallace, E.; Bridge, J.A.; Mayne, D.J.; Beqaj, S.; Sexton, W.J.; Lotan, Y.; Weizer, A.Z.; Jansz, G.K.; et al. Validation of an mRNA-based urine test for the detection of bladder cancer in patients with haematuria. Eur. Urol. Oncol. 2020. [Google Scholar] [CrossRef]
- Hurle, R.; Lazzeri, M.; Vanni, E.; Lughezzani, G.; Buffi, N.; Casale, P.; Saita, A.; Morenghi, E.; Forni, G.; Cardone, P.; et al. Active surveillance for low risk nonmuscle invasive bladder cancer: A confirmatory and resource consumption study from the BIAS project. J. Urol. 2018, 199, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, V.; Llorente, C.; de la Pena, E.; Perez-Fernandez, E.; Guijarro, A.; Sola, I. Long-term oncological outcomes of an active surveillance program in recurrent low grade Ta bladder cancer. Urol. Oncol. 2016, 34, 165.e119–165.e123. [Google Scholar] [CrossRef]
- D´Elia, C.; Pycha, A.; Folchini, D.M.; Mian, C.; Hanspeter, E.; Schwienbacher, C.; Vjaters, E.; Pycha, A.; Trenti, E. Diagnostic predictive value of Xpert Bladder Cancer Monitor in the follow-up of patients affected by non-muscle invasive bladder cancer. J. Clin. Pathol. 2018, 72, 140–144. [Google Scholar] [CrossRef]
- Pichler, R.; Fritz, J.; Tulchiner, G.; Klinglmair, G.; Soleiman, A.; Horninger, W.; Klocker, H.; Heidegger, I. Increased accuracy of a novel mRNA-based urine test for bladder cancer surveillance. BJU Int. 2018, 121, 29–37. [Google Scholar] [CrossRef] [Green Version]
- Valenberg, F.; Hiar, A.M.; Wallace, E.; Bridge, J.A.; Mayne, D.J.; Beqaj, S.; Sexton, W.J.; Lotan, Y.; Weizer, A.Z.; Jansz, G.K.; et al. Prospective validation of an mRNA-based urine test for surveillance of patients with bladder cancer. Eur. Urol. 2019, 75, 853–860. [Google Scholar] [CrossRef]
- Hurle, R.; Casale, P.; Saita, A.; Colombo, P.; Elefante, G.M.; Lughezzani, G.; Fasulo, V.; Paciotti, M.; Domanico, L.; Bevilacqua, G.; et al. Clinical performance of Xpert Bladder Cancer (BC) Monitor, a mRNA-based urine test, in active surveillance (AS) patients with recurrent non-muscle-invasive bladder cancer (NMIBC): Results from the Bladder Cancer Italian Active Surveillance (BIAS) project. World J. Urol. 2020, 38, 2215–2220. [Google Scholar] [CrossRef]
- Cappellen, D.; De Oliveira, C.; Ricol, D.; de Medina, S.; Bourdin, J.; Sastre-Garau, X.; Chopin, D.; Thiery, J.P.; Radvanyi, F. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat. Genet. 1999, 23, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Couffignal, C.; Desgrandchamps, F.; Mongiat-Artus, P.; Ravery, V.; Ouzaid, I.; Roupret, M.; Phe, V.; Ciofu, C.; Tubach, F.; Mentre, F.; et al. The diagnostic and prognostic performance of urinary FGFR3 mutation analysis in bladder cancer surveillance: A prospective multicenter study. Urology 2015, 86, 1185–1190. [Google Scholar] [CrossRef]
- Knowles, M.A. Role of FGFR3 in urothelial cell carcinoma: Biomarker and potential therapeutic target. World J. Urol. 2007, 25, 581–593. [Google Scholar] [CrossRef] [Green Version]
- Billerey, C.; Chopin, D.; Aubriot-Lorton, M.H.; Ricol, D.; Gil Diez de Medina, S.; Van Rhijn, B.; Bralet, M.P.; Lefrere-Belda, M.A.; Lahaye, J.B.; Abbou, C.C.; et al. Frequent FGFR3 mutations in papillary non-invasive bladder (pTa) tumors. Am. J. Pathol. 2001, 158, 1955–1959. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, Y.; Fujita, K.; Matsuzaki, K.; Eich, M.L.; Tomiyama, E.; Matsushita, M.; Koh, Y.; Nakano, K.; Wang, C.; Ishizuya, Y.; et al. Clinical significance of hotspot mutation analysis of urinary cell-free DNA in urothelial bladder cancer. Front. Oncol. 2020, 10, 755. [Google Scholar] [CrossRef]
- Chou, R.; Buckley, D.; Fu, R.; Gore, J.L.; Gustafson, K.; Griffin, J.; Grusing, S.; Selph, S. Emerging Approaches to Diagnosis and Treatment of Non-Muscle-Invasive Bladder Cancer; Agency for Healthcare Research and Quality: Rockville, MD, USA, 2015. [Google Scholar]
- Babjuk, M.; Bohle, A.; Burger, M.; Capoun, O.; Cohen, D.; Comperat, E.M.; Hernandez, V.; Kaasinen, E.; Palou, J.; Roupret, M.; et al. EAU Guidelines on non-muscle-invasive urothelial carcinoma of the bladder: Update 2016. Eur. Urol. 2017, 71, 447–461. [Google Scholar] [CrossRef]
- Lenherr, S.M.; Tsai, S.; Silva Neto, B.; Sullivan, T.B.; Cimmino, C.B.; Logvinenko, T.; Gee, J.; Huang, W.; Libertino, J.A.; Summerhayes, I.C.; et al. MicroRNA expression profile identifies high grade, non-muscle-invasive bladder tumors at elevated risk to progress to an invasive phenotype. Genes 2017, 8, 77. [Google Scholar] [CrossRef] [Green Version]
- Olivieri, M.; Ferro, M.; Terreri, S.; Durso, M.; Romanelli, A.; Avitabile, C.; De Cobelli, O.; Messere, A.; Bruzzese, D.; Vannini, I.; et al. Long non-coding RNA containing ultraconserved genomic region 8 promotes bladder cancer tumorigenesis. Oncotarget 2016, 7, 20636–20654. [Google Scholar] [CrossRef] [PubMed]
- Terreri, S.; Durso, M.; Colonna, V.; Romanelli, A.; Terracciano, D.; Ferro, M.; Perdona, S.; Castaldo, L.; Febbraio, F.; de Nigris, F.; et al. New cross-talk layer between ultraconserved non-coding RNAs, MicroRNAs and polycomb protein YY1 in bladder cancer. Genes 2016, 7, 127. [Google Scholar] [CrossRef] [PubMed]
- Pop-Bica, C.; Gulei, D.; Cojocneanu-Petric, R.; Braicu, C.; Petrut, B.; Berindan-Neagoe, I. Understanding the role of non-coding RNAs in bladder cancer: From dark matter to valuable therapeutic targets. Int. J. Mol. Sci. 2017, 18, 1514. [Google Scholar] [CrossRef] [PubMed]
- Seitz, A.K.; Christensen, L.L.; Christensen, E.; Faarkrog, K.; Ostenfeld, M.S.; Hedegaard, J.; Nordentoft, I.; Nielsen, M.M.; Palmfeldt, J.; Thomson, M.; et al. Profiling of long non-coding RNAs identifies LINC00958 and LINC01296 as candidate oncogenes in bladder cancer. Sci. Rep. 2017, 7, 395. [Google Scholar] [CrossRef] [Green Version]
- Terracciano, D.; Ferro, M.; Terreri, S.; Lucarelli, G.; D’Elia, C.; Musi, G.; de Cobelli, O.; Mirone, V.; Cimmino, A. Urinary long noncoding RNAs in nonmuscle-invasive bladder cancer: New architects in cancer prognostic biomarkers. Transl. Res. 2017, 184, 108–117. [Google Scholar] [CrossRef]
- Berindan-Neagoe, I.; Monroig Pdel, C.; Pasculli, B.; Calin, G.A. MicroRNAome genome: A treasure for cancer diagnosis and therapy. CA Cancer J. Clin. 2014, 64, 311–336. [Google Scholar] [CrossRef] [PubMed]
- Eissa, S.; Matboli, M.; Essawy, N.O.; Kotb, Y.M. Integrative functional genetic-epigenetic approach for selecting genes as urine biomarkers for bladder cancer diagnosis. Tumour Biol. 2015, 36, 9545–9552. [Google Scholar] [CrossRef]
- Breen, V.; Kasabov, N.; Kamat, A.M.; Jacobson, E.; Suttie, J.M.; O’Sullivan, P.J.; Kavalieris, L.; Darling, D.G. A holistic comparative analysis of diagnostic tests for urothelial carcinoma: A study of Cxbladder Detect, UroVysion(R) FISH, NMP22(R) and cytology based on imputation of multiple datasets. BMC Med. Res. Methodol. 2015, 15, 45. [Google Scholar] [CrossRef] [Green Version]
- Zuiverloon, T.C.; Nieuweboer, A.J.; Vekony, H.; Kirkels, W.J.; Bangma, C.H.; Zwarthoff, E.C. Markers predicting response to bacillus Calmette-Guerin immunotherapy in high-risk bladder cancer patients: A systematic review. Eur. Urol. 2012, 61, 128–145. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.M.; Li, H.X. Tumor heterogeneity and the potential role of liquid biopsy in bladder cancer. Cancer Commun. 2020. [Google Scholar] [CrossRef]
- Kamat, A.M.; Briggman, J.; Urbauer, D.L.; Svatek, R.; Nogueras Gonzalez, G.M.; Anderson, R.; Grossman, H.B.; Prat, F.; Dinney, C.P. Cytokine panel for response to Intravesical therapy (CyPRIT): Nomogram of changes in urinary cytokine levels predicts patient response to bacillus calmette-guerin. Eur. Urol. 2016, 69, 197–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmasi, A.; Elashoff, D.A.; Guo, R.; Upfill-Brown, A.; Rosser, C.J.; Rose, J.M.; Giffin, L.C.; Gonzalez, L.E.; Chamie, K. Urinary cytokine profile to predict response to intravesical BCG with or without HS-410 therapy in patients with non-muscle-invasive bladder cancer. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1036–1044. [Google Scholar] [CrossRef] [PubMed]
- Franovic, A.; Raymond, V.M.; Erlander, M.G.; Reckamp, K.L. Urine test for EGFR analysis in patients with non-small cell lung cancer. J. Thorac Dis. 2017, 9, S1323–S1331. [Google Scholar] [CrossRef] [Green Version]
- Choudhury, N.J.; Campanile, A.; Antic, T.; Yap, K.L.; Fitzpatrick, C.A.; Wade, J.L., 3rd; Karrison, T.; Stadler, W.M.; Nakamura, Y.; O’Donnell, P.H. Afatinib activity in platinum-refractory metastatic urothelial carcinoma in patients with ERBB alterations. J. Clin. Oncol. 2016, 34, 2165–2171. [Google Scholar] [CrossRef] [Green Version]
- Messing, E. Markers of detection. Urol. Oncol. 2007, 25, 344–347. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.C.; Chang, S.S.; Dalbagni, G.; Pruthi, R.S.; Seigne, J.D.; Skinner, E.C.; Wolf, J.S., Jr.; Schellhammer, P.F. Guideline for the management of nonmuscle invasive bladder cancer (stages Ta, T1, and Tis): 2007 update. J. Urol. 2007, 178, 2314–2330. [Google Scholar] [CrossRef]
- Shariat, S.F.; Karam, J.A.; Lotan, Y.; Karakiewizc, P.I. Critical evaluation of urinary markers for bladder cancer detection and monitoring. Rev. Urol. 2008, 10, 120–135. [Google Scholar] [PubMed]
- Ponsky, L.E.; Sharma, S.; Pandrangi, L.; Kedia, S.; Nelson, D.; Agarwal, A.; Zippe, C.D. Screening and monitoring for bladder cancer: Refining the use of NMP22. J. Urol. 2001, 166, 75–78. [Google Scholar] [CrossRef]
- Thomas, L.; Leyh, H.; Marberger, M.; Bombardieri, E.; Bassi, P.; Pagano, F.; Pansadoro, V.; Sternberg, C.N.; Boccon-Gibod, L.; Ravery, V.; et al. Multicenter trial of the quantitative BTA TRAK assay in the detection of bladder cancer. Clin. Chem. 1999, 45, 472–477. [Google Scholar]
- Goodison, S.; Ogawa, O.; Matsui, Y.; Kobayashi, T.; Miyake, M.; Ohnishi, S.; Fujimoto, K.; Dai, Y.; Shimizu, Y.; Tsukikawa, K.; et al. A multiplex urinary immunoassay for bladder cancer detection: Analysis of a Japanese cohort. J. Transl. Med. 2016, 14, 287. [Google Scholar] [CrossRef] [Green Version]
- Chung, W.; Bondaruk, J.; Jelinek, J.; Lotan, Y.; Liang, S.; Czerniak, B.; Issa, J.P. Detection of bladder cancer using novel DNA methylation biomarkers in urine sediments. Cancer Epidemiol. Biomark. Prev. 2011, 20, 1483–1491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mengual, L.; Burset, M.; Ribal, M.J.; Ars, E.; Marin-Aguilera, M.; Fernandez, M.; Ingelmo-Torres, M.; Villavicencio, H.; Alcaraz, A. Gene expression signature in urine for diagnosing and assessing aggressiveness of bladder urothelial carcinoma. Clin. Cancer Res. 2010, 16, 2624–2633. [Google Scholar] [CrossRef] [Green Version]
- Holyoake, A.; O’Sullivan, P.; Pollock, R.; Best, T.; Watanabe, J.; Kajita, Y.; Matsui, Y.; Ito, M.; Nishiyama, H.; Kerr, N.; et al. Development of a multiplex RNA urine test for the detection and stratification of transitional cell carcinoma of the bladder. Clin. Cancer Res. 2008, 14, 742–749. [Google Scholar] [CrossRef] [Green Version]
- Yeung, C.; Dinh, T.; Lee, J. The health economics of bladder cancer: An updated review of the published literature. Pharmacoeconomics 2014, 32, 1093–1104. [Google Scholar] [CrossRef]
- Meeks, J.J.; Al-Ahmadie, H.; Faltas, B.M.; Taylor, J.A., 3rd; Flaig, T.W.; DeGraff, D.J.; Christensen, E.; Woolbright, B.L.; McConkey, D.J.; Dyrskjot, L. Genomic heterogeneity in bladder cancer: Challenges and possible solutions to improve outcomes. Nat. Rev. Urol. 2020, 17, 259–270. [Google Scholar] [CrossRef]
- Warrick, J.I.; Sjodahl, G.; Kaag, M.; Raman, J.D.; Merrill, S.; Shuman, L.; Chen, G.; Walter, V.; DeGraff, D.J. Intratumoral heterogeneity of bladder cancer by molecular subtypes and histologic variants. Eur. Urol. 2019, 75, 18–22. [Google Scholar] [CrossRef]

| Test | Variables | Method | Use in Clinical Practice | Cost | Approval | Reference |
|---|---|---|---|---|---|---|
| Bladder EpiCheck | DNA methylation pattern (15 genomic biomarkers) | RT-PCR | Diagnosis | NA | Yes (FDA/CE) | [27,28,29,30,31,32] |
| Urodiag | FGFR3 mutations and epigenetic markers (hypermethylation of HS3ST2, SEPTIN9 and SLIT2) | PCR | Diagnosis | NA | Yes (CE) | [33,34,35] |
| AssureMDX | FGFR3, TERT, HRAS mutations, methylation of OTX1, ONECUT2, TWIST1 | PCR | Diagnosis | NA | No | [36,37] |
| UroMuTERT | TERT promoter mutations (C228T and C250T) | PCR | Diagnosis | NA | No | [38,39,40,41,42] |
| Uromonitor-V2 | FGFR3, TERT and KRAS hotspot mutations | qPCR | Detection of NMIBC recurrence | NA | Yes (FDA/CE) | [43,44] |
| Urovysion | 3,7,17 chromosome aneuploidy and loss of 9p21 locus | FISH | Diagnosis | 200$/test | Yes (FDA/CE) | [45,46,47,48,49] |
| Uroseek® | Mutations in 11 genes and aneuploidy | NGS/SANGER | Diagnosis | 1000$/test | No | [50,51] |
| uCAPP-Seq | Urine ctDNA determination | NGS | Diagnosis | NA | No | [52] |
| NMP22 | Nuclear matrix protein released by dead cells in urine | ELISA | Surveillance in combination with cystoscopy | 25$/test | Yes (FDA/CE) | [53,54,55] |
| BTA | Basement membrane-derived antigen released by cancer cells | ELISA | Surveillance in combination with cystoscopy | 40$/test | Yes (FDA/CE) | [56,57,58,59] |
| ADXBLADDER | MCM5 protein | ELISA | Surveillance; prognosis | 52$/test | Yes (CE) | [60,61,62,63,64,65,66,67] |
| Oncuria | 10 protein biomarkers (APOE, ANG, A1AT, CA9, IL8, MMP9, MMP10, PAI1, SDC1, VEGFA) | ELISA | Diagnosis | NA | No | [68] |
| miRNAs | miR-200, miR-145, and miR-214 | RT-qPCR | Prognosis | NA | No | [69,70,71] |
| LncRNA | UCA-1, UCA1-203, UCA1-201, MALAT1 and LINC00355 | RT-qPCR | Diagnosis | NA | No | [72,73,74,75] |
| Cxbladder detect and monitor | mRNA signature in urine (CDK1, MDK, HOXA13, IGFBP5 and CXCR2) | RT-qPCR | Diagnosis, surveillance | NA | Yes (CE) | [76,77,78,79] |
| Xpert Bladder cancer detect and monitor | mRNA signature in urine (ABL1, ANXA10, CRH, IGF2 and UPK1B) | RT-qPCR | Diagnosis, surveillance | 165$/test | Yes (CE) | [80,81,82,83,84,85,86,87] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferro, M.; La Civita, E.; Liotti, A.; Cennamo, M.; Tortora, F.; Buonerba, C.; Crocetto, F.; Lucarelli, G.; Busetto, G.M.; Del Giudice, F.; et al. Liquid Biopsy Biomarkers in Urine: A Route towards Molecular Diagnosis and Personalized Medicine of Bladder Cancer. J. Pers. Med. 2021, 11, 237. https://doi.org/10.3390/jpm11030237
Ferro M, La Civita E, Liotti A, Cennamo M, Tortora F, Buonerba C, Crocetto F, Lucarelli G, Busetto GM, Del Giudice F, et al. Liquid Biopsy Biomarkers in Urine: A Route towards Molecular Diagnosis and Personalized Medicine of Bladder Cancer. Journal of Personalized Medicine. 2021; 11(3):237. https://doi.org/10.3390/jpm11030237
Chicago/Turabian StyleFerro, Matteo, Evelina La Civita, Antonietta Liotti, Michele Cennamo, Fabiana Tortora, Carlo Buonerba, Felice Crocetto, Giuseppe Lucarelli, Gian Maria Busetto, Francesco Del Giudice, and et al. 2021. "Liquid Biopsy Biomarkers in Urine: A Route towards Molecular Diagnosis and Personalized Medicine of Bladder Cancer" Journal of Personalized Medicine 11, no. 3: 237. https://doi.org/10.3390/jpm11030237