Proteomics of Extracellular Vesicles: Update on Their Composition, Biological Roles and Potential Use as Diagnostic Tools in Atherosclerotic Cardiovascular Diseases
Abstract
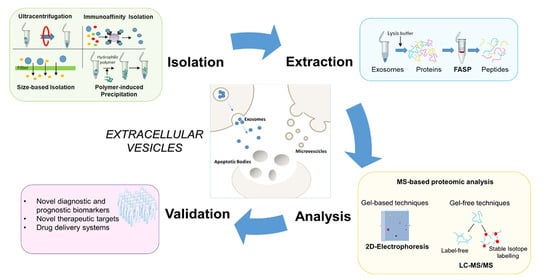
:1. Introduction
1.1. Exosomes
1.2. Microvesicles
1.3. Apoptotic Bodies
2. Emerging Role of Extracellular Vesicles
3. Proteomic Methods
3.1. Sample Collection, Storage and Processing
3.2. Isolation Methods for MS-Based Proteomic Studies of Extracellular Vesicles
3.2.1. Differential Ultracentrifugation
3.2.2. Density-Gradient Ultracentrifugation
3.2.3. Size-Based Isolation
3.2.4. Immunoaffinity Isolation
3.2.5. Polymer-Induced Precipitation
3.3. MS-Based Strategies for the Study of Extracellular Vesicle Proteome
4. Exosomes in Cardiovascular Diseases
4.1. Exosomes Derived from Cardiomyocytes
4.2. Exosomes Derived from Mesenchymal Stem Cell
4.3. Exosomes Derived from Cardiac Fibroblasts
4.4. Exosomes Derived from Endothelial Cells
4.5. Exosomes Derived from Vascular Smooth Muscle Cells
4.6. Exosomes Derived from Cardiac-Derived Progenitor Cells
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 2-DE | Two-dimensional gel electrophoresis |
| ACR | Acute cellular reaction |
| AF4 | Asymmetric flow field-flow fractionation |
| Akt | Protein kinase B |
| ALIX | Programmed cell death 6-interacting protein |
| AMI | Acute myocardial infarction |
| AMPK | AMP-activated protein kinase |
| AMR | Antibody-mediated reaction |
| Ang II | Angiotensin II |
| AT1R | Ang II receptor type 1 |
| AT2R | Ang II receptor type 1 |
| BMC | Bone marrow-derived mesenchymal stem/progenitor cell |
| CAA | Coronary artery aneurism |
| CAD | Coronary artery disease |
| CDC | Cardiosphere-derived cell |
| CF | Cardiac fibroblast |
| CHAPS | 3-((3-cholamidopropyl)dimethylammonio)-1-propanesulfonic |
| c-JNK | c-Jun N-terminal kinase |
| CPC | Cardiac-derived progenitor cell |
| CVD | Cardiovascular disease |
| DDA | Data dependent acquisition |
| DIA | Data-independent acquisition |
| Dll4 | Delta-like 4 |
| DTT | Dithiothreitol |
| DU | De-ubiquitylating enzymes |
| EC | Endothelial cell |
| ECM | Extracellular matrix |
| EMT | Epithelial-to-mesenchymal transition |
| ERK | Extracellular signal-regulated kinase |
| ESCRT | Endosomal sorting complex required |
| EV | Extracellular vesicle |
| FASP | Filter aided sample preparation approach |
| FFF | Field flow fractionation |
| GPCR | G protein-coupled receptor |
| GPI | Glycosylphosphatidylinositol |
| HIF-1α | Hypoxia inducible factor 1α |
| HRS | Hepatocyte growth factor regulated tyrosine kinase substrate |
| HSC | Heat shock cognate |
| HSP | Heat shock protein |
| IGF1 | Insulin-like growth factor 1 |
| IGFBP-4 | Insulin-like growth factor binding protein 4 |
| IL-1β | Interleukin 1β |
| ILV | Intraluminal vesicle |
| IMAC | Immobilized metal ion affinity chromatography |
| iTRAQ | Isobaric tags for relative and absolute quantitation |
| IVIG | Intravenous dose of immunoglobulin |
| KD | Kawasaki disease |
| LAMP 1/2 | Lysosomal associated membrane protein 1/2 |
| LC | Liquid chromatography |
| MAPK | Mitogen-activated protein kinase |
| MED | Multiple enzyme digestion |
| MI | Myocardial Infarction |
| MRM | Multiple reaction monitoring |
| MS | Mass spectrometry |
| MSC | Mesenchymal stem cell |
| MV | Microvesicle |
| MVB | Multivesicular body |
| NFkB | Nuclear factor-kappa B |
| PAD | Peripheral arterial disease |
| PAPP-A | Pregnancy associated plasma protein-A |
| PAR1 | Protease-activated receptor 1 |
| PBS | Phosphate buffered saline |
| PCA | Principal component analysis |
| PDGF-D | Platelet derived growth factor D |
| PDGFR-β | Platelet derived growth factor receptor β |
| PEG | Polyethylene glycol |
| PI3K | Phosphatidylinositol 3-kinase |
| PRM-MS | Parallel reaction monitoring-mass spectrometry |
| PROMIS-Quan | Proteomics of microparticles using super-SILAC quantification |
| PT | Prothrombin |
| PUC-ERLIC | Prolonged ultracentrifugation-electrostatic repulsion-hydrophilic interaction chromatography |
| RAS | Renin angiotensin system |
| ROCK | RHO associated protein kinase |
| ROS | Reactive oxygen species |
| SDS | Sodium dodecyl sulphate |
| SEC | Size exclusion chromatography |
| SILAC | Stable isotope labelling with amino acids in cell culture |
| SNARE | Soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor |
| SOD-1 | Superoxide dismutase 1 |
| STAM | Signal-transducing adaptor molecule |
| SWATH | Sequential window acquisition of all theoretical fragment ion |
| TFF | Tangential flow filtration |
| TiO2 | Titanium dioxide |
| TLR | Toll like receptor |
| TNF-α | Tissue necrosis factor α |
| VPS4 | Vacuolar protein sorting-associated protein 4 |
| VSMC | Vascular smooth muscle cell |
| VTA1 | Vacuolar protein sorting-associated protein VTA1 homolog |
References
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, R.; Patel, T.; Freedman, J.E. Circulating Extracellular Vesicles in Human Disease. N. Engl. J. Med. 2018, 379, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neurooncol. 2013, 113, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pieragostino, D.; Lanuti, P.; Cicalini, I.; Cufaro, M.C.; Ciccocioppo, F.; Ronci, M.; Simeone, P.; Onofrj, M.; van der Pol, E.; Fontana, A.; et al. Proteomics characterization of extracellular vesicles sorted by flow cytometry reveals a disease-specific molecular cross-talk from cerebrospinal fluid and tears in multiple sclerosis. J. Proteom. 2019, 204, 103403. [Google Scholar] [CrossRef] [PubMed]
- Al-Nedawi, K.; Meehan, B.; Rak, J. Microvesicles: Messengers and mediators of tumor progression. Cell Cycle 2009, 8, 2014–2018. [Google Scholar] [CrossRef]
- Brennan, K.; Martin, K.; FitzGerald, S.P.; O’Sullivan, J.; Wu, Y.; Blanco, A.; Richardson, C.; Mc Gee, M.M. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci. Rep. 2020, 10, 1039. [Google Scholar] [CrossRef] [Green Version]
- Konoshenko, M.Y.; Lekchnov, E.A.; Vlassov, A.V.; Laktionov, P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed. Res. Int. 2018, 2018, 8545347. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Pan, B.T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Yoon, Y.J.; Kim, O.Y.; Gho, Y.S. Extracellular vesicles as emerging intercellular communicasomes. BMB Rep. 2014, 47, 531–539. [Google Scholar] [CrossRef]
- Tancini, B.; Buratta, S.; Sagini, K.; Costanzi, E.; Delo, F.; Urbanelli, L.; Emiliani, C. Insight into the Role of Extracellular Vesicles in Lysosomal Storage Disorders. Genes 2019, 10, 510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eitan, E.; Suire, C.; Zhang, S.; Mattson, M.P. Impact of lysosome status on extracellular vesicle content and release. Ageing Res. Rev. 2016, 32, 65–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buratta, S.; Tancini, B.; Sagini, K.; Delo, F.; Chiaradia, E.; Urbanelli, L.; Emiliani, C. Lysosomal Exocytosis, Exosome Release and Secretory Autophagy: The Autophagic- and Endo-Lysosomal Systems Go Extracellular. Int. J. Mol. Sci. 2020, 21, 2576. [Google Scholar] [CrossRef] [Green Version]
- De Gassart, A.; Geminard, C.; Fevrier, B.; Raposo, G.; Vidal, M. Lipid raft-associated protein sorting in exosomes. Blood 2003, 102, 4336–4344. [Google Scholar] [CrossRef] [Green Version]
- Thery, C.; Boussac, M.; Veron, P.; Ricciardi-Castagnoli, P.; Raposo, G.; Garin, J.; Amigorena, S. Proteomic analysis of dendritic cell-derived exosomes: A secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 2001, 166, 7309–7318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mateescu, B.; Kowal, E.J.; van Balkom, B.W.; Bartel, S.; Bhattacharyya, S.N.; Buzas, E.I.; Buck, A.H.; de Candia, P.; Chow, F.W.; Das, S.; et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA—An ISEV position paper. J. Extracell. Vesicles 2017, 6, 1286095. [Google Scholar] [CrossRef] [Green Version]
- Henne, W.M.; Buchkovich, N.J.; Emr, S.D. The ESCRT pathway. Dev. Cell 2011, 21, 77–91. [Google Scholar] [CrossRef] [Green Version]
- McGough, I.J.; Vincent, J.P. Exosomes in developmental signalling. Development 2016, 143, 2482–2493. [Google Scholar] [CrossRef] [Green Version]
- Hurley, J.H. The ESCRT complexes. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 463–487. [Google Scholar] [CrossRef]
- Dores, M.R.; Trejo, J. Ubiquitination of G protein-coupled receptors: Functional implications and drug discovery. Mol. Pharmacol. 2012, 82, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brugger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Stuffers, S.; Brech, A.; Stenmark, H. ESCRT proteins in physiology and disease. Exp. Cell Res. 2009, 315, 1619–1626. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell 2011, 21, 708–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kajimoto, T.; Okada, T.; Miya, S.; Zhang, L.; Nakamura, S. Ongoing activation of sphingosine 1-phosphate receptors mediates maturation of exosomal multivesicular endosomes. Nat. Commun. 2013, 4, 2712. [Google Scholar] [CrossRef] [Green Version]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [Green Version]
- Greening, D.W.; Simpson, R.J. Understanding extracellular vesicle diversity—Current status. Expert Rev. Proteom. 2018, 15, 887–910. [Google Scholar] [CrossRef]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef]
- Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Borges, F.T.; Reis, L.A.; Schor, N. Extracellular vesicles: Structure, function, and potential clinical uses in renal diseases. Braz. J. Med. Biol. Res. 2013, 46, 824–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piccin, A.; Murphy, W.G.; Smith, O.P. Circulating microparticles: Pathophysiology and clinical implications. Blood Rev. 2007, 21, 157–171. [Google Scholar] [CrossRef]
- Antonyak, M.A.; Cerione, R.A. Microvesicles as mediators of intercellular communication in cancer. Methods Mol. Biol. 2014, 1165, 147–173. [Google Scholar] [CrossRef] [PubMed]
- Bolukbasi, M.F.; Mizrak, A.; Ozdener, G.B.; Madlener, S.; Strobel, T.; Erkan, E.P.; Fan, J.B.; Breakefield, X.O.; Saydam, O. miR-1289 and “Zipcode”-like Sequence Enrich mRNAs in Microvesicles. Mol. Ther. Nucleic Acids 2012, 1, e10. [Google Scholar] [CrossRef]
- Kerr, J.F.; Wyllie, A.H.; Currie, A.R. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef] [Green Version]
- Gyorgy, B.; Szabo, T.G.; Pasztoi, M.; Pal, Z.; Misjak, P.; Aradi, B.; Laszlo, V.; Pallinger, E.; Pap, E.; Kittel, A.; et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Cell Mol. Life Sci. 2011, 68, 2667–2688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedlund, M.; Nagaeva, O.; Kargl, D.; Baranov, V.; Mincheva-Nilsson, L. Thermal- and oxidative stress causes enhanced release of NKG2D ligand-bearing immunosuppressive exosomes in leukemia/lymphoma T and B cells. PLoS ONE 2011, 6, e16899. [Google Scholar] [CrossRef]
- De Jong, O.G.; Verhaar, M.C.; Chen, Y.; Vader, P.; Gremmels, H.; Posthuma, G.; Schiffelers, R.M.; Gucek, M.; van Balkom, B.W. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J. Extracell. Vesicles 2012, 1. [Google Scholar] [CrossRef]
- Boulanger, C.M.; Loyer, X.; Rautou, P.E.; Amabile, N. Extracellular vesicles in coronary artery disease. Nat. Rev. Cardiol. 2017, 14, 259–272. [Google Scholar] [CrossRef]
- Amabile, N.; Cheng, S.; Renard, J.M.; Larson, M.G.; Ghorbani, A.; McCabe, E.; Griffin, G.; Guerin, C.; Ho, J.E.; Shaw, S.Y.; et al. Association of circulating endothelial microparticles with cardiometabolic risk factors in the Framingham Heart Study. Eur. Heart J. 2014, 35, 2972–2979. [Google Scholar] [CrossRef] [PubMed]
- Conde-Vancells, J.; Gonzalez, E.; Lu, S.C.; Mato, J.M.; Falcon-Perez, J.M. Overview of extracellular microvesicles in drug metabolism. Expert Opin. Drug Metab. Toxicol. 2010, 6, 543–554. [Google Scholar] [CrossRef] [Green Version]
- Rowland, A.; Ruanglertboon, W.; van Dyk, M.; Wijayakumara, D.; Wood, L.S.; Meech, R.; Mackenzie, P.I.; Rodrigues, A.D.; Marshall, J.C.; Sorich, M.J. Plasma extracellular nanovesicle (exosome)-derived biomarkers for drug metabolism pathways: A novel approach to characterize variability in drug exposure. Br. J. Clin. Pharmacol. 2019, 85, 216–226. [Google Scholar] [CrossRef] [Green Version]
- Tickner, J.A.; Urquhart, A.J.; Stephenson, S.A.; Richard, D.J.; O’Byrne, K.J. Functions and therapeutic roles of exosomes in cancer. Front. Oncol. 2014, 4, 127. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panfoli, I.; Santucci, L.; Bruschi, M.; Petretto, A.; Calzia, D.; Ramenghi, L.A.; Ghiggeri, G.; Candiano, G. Microvesicles as promising biological tools for diagnosis and therapy. Expert Rev. Proteom. 2018, 15, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef]
- Bobrie, A.; Krumeich, S.; Reyal, F.; Recchi, C.; Moita, L.F.; Seabra, M.C.; Ostrowski, M.; Thery, C. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012, 72, 4920–4930. [Google Scholar] [CrossRef] [Green Version]
- Samir, E.L.A.; Mager, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- Catalano, M.; O’Driscoll, L. Inhibiting extracellular vesicles formation and release: A review of EV inhibitors. J. Extracell. Vesicles 2020, 9, 1703244. [Google Scholar] [CrossRef] [Green Version]
- Dinkins, M.B.; Dasgupta, S.; Wang, G.; Zhu, G.; Bieberich, E. Exosome reduction in vivo is associated with lower amyloid plaque load in the 5XFAD mouse model of Alzheimer’s disease. Neurobiol. Aging 2014, 35, 1792–1800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaput, N.; Thery, C. Exosomes: Immune properties and potential clinical implementations. Semin. Immunopathol. 2011, 33, 419–440. [Google Scholar] [CrossRef] [PubMed]
- Arslan, F.; Lai, R.C.; Smeets, M.B.; Akeroyd, L.; Choo, A.; Aguor, E.N.; Timmers, L.; van Rijen, H.V.; Doevendans, P.A.; Pasterkamp, G.; et al. Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res. 2013, 10, 301–312. [Google Scholar] [CrossRef] [Green Version]
- Ophelders, D.R.; Wolfs, T.G.; Jellema, R.K.; Zwanenburg, A.; Andriessen, P.; Delhaas, T.; Ludwig, A.K.; Radtke, S.; Peters, V.; Janssen, L.; et al. Mesenchymal Stromal Cell-Derived Extracellular Vesicles Protect the Fetal Brain After Hypoxia-Ischemia. Stem Cells Transl. Med. 2016, 5, 754–763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballini, A.; Cantore, S.; Scacco, S.; Coletti, D.; Tatullo, M. Mesenchymal Stem Cells as Promoters, Enhancers, and Playmakers of the Translational Regenerative Medicine 2018. Stem Cells Int. 2018, 2018, 6927401. [Google Scholar] [CrossRef] [Green Version]
- Ballini, A.; Scacco, S.; Coletti, D.; Pluchino, S.; Tatullo, M. Mesenchymal Stem Cells as Promoters, Enhancers, and Playmakers of the Translational Regenerative Medicine. Stem Cells Int. 2017, 2017, 3292810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Shen, L.; Shi, H.; Pan, Z.; Wu, L.; Yan, Y.; Zhang, X.; Mao, F.; Qian, H.; Xu, W. Exosomes from Human Umbilical Cord Mesenchymal Stem Cells: Identification, Purification, and Biological Characteristics. Stem Cells Int. 2016, 2016, 1929536. [Google Scholar] [CrossRef]
- Ma, T.; Chen, Y.; Chen, Y.; Meng, Q.; Sun, J.; Shao, L.; Yu, Y.; Huang, H.; Hu, Y.; Yang, Z.; et al. MicroRNA-132, Delivered by Mesenchymal Stem Cell-Derived Exosomes, Promote Angiogenesis in Myocardial Infarction. Stem Cells Int. 2018, 2018, 3290372. [Google Scholar] [CrossRef] [Green Version]
- Spagnuolo, G.; Codispoti, B.; Marrelli, M.; Rengo, C.; Rengo, S.; Tatullo, M. Commitment of Oral-Derived Stem Cells in Dental and Maxillofacial Applications. Dent. J. 2018, 6, 72. [Google Scholar] [CrossRef] [Green Version]
- Govindasamy, V.; Ronald, V.S.; Abdullah, A.N.; Ganesan Nathan, K.R.; Aziz, Z.A.; Abdullah, M.; Zain, R.B.; Kasim, N.H.; Musa, S.; Bhonde, R.R. Human platelet lysate permits scale-up of dental pulp stromal cells for clinical applications. Cytotherapy 2011, 13, 1221–1233. [Google Scholar] [CrossRef] [Green Version]
- Marrazzo, P.; Paduano, F.; Palmieri, F.; Marrelli, M.; Tatullo, M. Highly Efficient In Vitro Reparative Behaviour of Dental Pulp Stem Cells Cultured with Standardised Platelet Lysate Supplementation. Stem Cells Int. 2016, 2016, 7230987. [Google Scholar] [CrossRef] [Green Version]
- Jang, S.C.; Gho, Y.S. Could bioengineered exosome-mimetic nanovesicles be an efficient strategy for the delivery of chemotherapeutics? Nanomedicine 2014, 9, 177–180. [Google Scholar] [CrossRef]
- Purushothaman, A. Exosomes from Cell Culture-Conditioned Medium: Isolation by Ultracentrifugation and Characterization. Methods Mol. Biol. 2019, 1952, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Sinha, A.; Principe, S.; Alfaro, J.; Ignatchenko, A.; Ignatchenko, V.; Kislinger, T. Proteomic Profiling of Secreted Proteins, Exosomes, and Microvesicles in Cell Culture Conditioned Media. Methods Mol. Biol. 2018, 1722, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, K.; Yan, I.K.; Patel, T. Isolation of Tissue Extracellular Vesicles from the Liver. J. Vis. Exp. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witwer, K.W.; Buzas, E.I.; Bemis, L.T.; Bora, A.; Lasser, C.; Lotvall, J.; Nolte-’t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef]
- Yuana, Y.; Boing, A.N.; Grootemaat, A.E.; van der Pol, E.; Hau, C.M.; Cizmar, P.; Buhr, E.; Sturk, A.; Nieuwland, R. Handling and storage of human body fluids for analysis of extracellular vesicles. J. Extracell. Vesicles 2015, 4, 29260. [Google Scholar] [CrossRef]
- Lacroix, R.; Judicone, C.; Poncelet, P.; Robert, S.; Arnaud, L.; Sampol, J.; Dignat-George, F. Impact of pre-analytical parameters on the measurement of circulating microparticles: Towards standardization of protocol. J. Thromb. Haemost. 2012, 10, 437–446. [Google Scholar] [CrossRef]
- Choi, J.Y.; Kim, S.; Kwak, H.B.; Park, D.H.; Park, J.H.; Ryu, J.S.; Park, C.S.; Kang, J.H. Extracellular Vesicles as a Source of Urological Biomarkers: Lessons Learned From Advances and Challenges in Clinical Applications to Major Diseases. Int. Neurourol. J. 2017, 21, 83–96. [Google Scholar] [CrossRef]
- Bari, E.; Perteghella, S.; Di Silvestre, D.; Sorlini, M.; Catenacci, L.; Sorrenti, M.; Marrubini, G.; Rossi, R.; Tripodo, G.; Mauri, P.; et al. Pilot Production of Mesenchymal Stem/Stromal Freeze-Dried Secretome for Cell-Free Regenerative Nanomedicine: A Validated GMP-Compliant Process. Cells 2018, 7, 190. [Google Scholar] [CrossRef] [Green Version]
- Bahr, M.M.; Amer, M.S.; Abo-El-Sooud, K.; Abdallah, A.N.; El-Tookhy, O.S. Preservation techniques of stem cells extracellular vesicles: A gate for manufacturing of clinical grade therapeutic extracellular vesicles and long-term clinical trials. Int. J. Vet. Sci. Med. 2020, 8, 1–8. [Google Scholar] [CrossRef]
- Bosch, S.; de Beaurepaire, L.; Allard, M.; Mosser, M.; Heichette, C.; Chretien, D.; Jegou, D.; Bach, J.M. Trehalose prevents aggregation of exosomes and cryodamage. Sci. Rep. 2016, 6, 36162. [Google Scholar] [CrossRef] [Green Version]
- Frank, J.; Richter, M.; de Rossi, C.; Lehr, C.M.; Fuhrmann, K.; Fuhrmann, G. Extracellular vesicles protect glucuronidase model enzymes during freeze-drying. Sci. Rep. 2018, 8, 12377. [Google Scholar] [CrossRef] [Green Version]
- Isacco, C.G.; Ballini, A.; De Vito, D.; Nguyen, K.C.D.; Cantore, S.; Bottalico, L.; Quagliuolo, L.; Boccellino, M.; Di Domenico, M.; Santacroce, L.; et al. Rebalance the oral microbiota as efficacy tool in endocrine, metabolic, and immune disorders. Endocr. Metab. Immune Disord. Drug Targets 2020. [Google Scholar] [CrossRef] [PubMed]
- Brahmer, A.; Neuberger, E.; Esch-Heisser, L.; Haller, N.; Jorgensen, M.M.; Baek, R.; Mobius, W.; Simon, P.; Kramer-Albers, E.M. Platelets, endothelial cells and leukocytes contribute to the exercise-triggered release of extracellular vesicles into the circulation. J. Extracell. Vesicles 2019, 8, 1615820. [Google Scholar] [CrossRef] [PubMed]
- Aharon, A.; Brenner, B. Microparticles and pregnancy complications. Thromb. Res. 2011, 127 (Suppl. S3), S67–S71. [Google Scholar] [CrossRef]
- Strohacker, K.; Breslin, W.L.; Carpenter, K.C.; Davidson, T.R.; Agha, N.H.; McFarlin, B.K. Moderate-intensity, premeal cycling blunts postprandial increases in monocyte cell surface CD18 and CD11a and endothelial microparticles following a high-fat meal in young adults. Appl. Physiol. Nutr. Metab. 2012, 37, 530–539. [Google Scholar] [CrossRef]
- Li, J.; He, X.; Deng, Y.; Yang, C. An Update on Isolation Methods for Proteomic Studies of Extracellular Vesicles in Biofluids. Molecules 2019, 24, 3516. [Google Scholar] [CrossRef] [Green Version]
- Momen-Heravi, F.; Balaj, L.; Alian, S.; Trachtenberg, A.J.; Hochberg, F.H.; Skog, J.; Kuo, W.P. Impact of biofluid viscosity on size and sedimentation efficiency of the isolated microvesicles. Front. Physiol. 2012, 3, 162. [Google Scholar] [CrossRef] [Green Version]
- Szatanek, R.; Baran, J.; Siedlar, M.; Baj-Krzyworzeka, M. Isolation of extracellular vesicles: Determining the correct approach (Review). Int. J. Mol. Med. 2015, 36, 11–17. [Google Scholar] [CrossRef] [Green Version]
- Chutipongtanate, S.; Greis, K.D. Multiplex Biomarker Screening Assay for Urinary Extracellular Vesicles Study: A Targeted Label-Free Proteomic Approach. Sci. Rep. 2018, 8, 15039. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.J.; Jin, D.D.; Jiang, F.; Liu, J.X.; Qu, L.S.; Ni, W.K.; Liu, Z.X.; Lu, C.H.; Ni, R.Z.; Zhu, J.; et al. Characterization and proteomic profiling of pancreatic cancer-derived serum exosomes. J. Cell Biochem. 2019, 120, 988–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, Z.; Pang, R.T.K.; Liu, W.; Li, Q.; Cheng, R.; Yeung, W.S.B. Polymer-based precipitation preserves biological activities of extracellular vesicles from an endometrial cell line. PLoS ONE 2017, 12, e0186534. [Google Scholar] [CrossRef] [Green Version]
- Shelke, G.V.; Lasser, C.; Gho, Y.S.; Lotvall, J. Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J. Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef] [Green Version]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3.22.1–3.22.29. [Google Scholar] [CrossRef]
- Muraoka, S.; Lin, W.; Chen, M.; Hersh, S.W.; Emili, A.; Xia, W.; Ikezu, T. Assessment of separation methods for extracellular vesicles from human and mouse brain tissues and human cerebrospinal fluids. Methods 2020, 177, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Dhondt, B.; Geeurickx, E.; Tulkens, J.; Van Deun, J.; Vergauwen, G.; Lippens, L.; Miinalainen, I.; Rappu, P.; Heino, J.; Ost, P.; et al. Unravelling the proteomic landscape of extracellular vesicles in prostate cancer by density-based fractionation of urine. J. Extracell. Vesicles 2020, 9, 1736935. [Google Scholar] [CrossRef]
- Kalra, H.; Adda, C.G.; Liem, M.; Ang, C.S.; Mechler, A.; Simpson, R.J.; Hulett, M.D.; Mathivanan, S. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics 2013, 13, 3354–3364. [Google Scholar] [CrossRef]
- Greening, D.W.; Xu, R.; Ji, H.; Tauro, B.J.; Simpson, R.J. A protocol for exosome isolation and characterization: Evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol. Biol. 2015, 1295, 179–209. [Google Scholar] [CrossRef]
- Wang, W.; Luo, J.; Wang, S. Recent Progress in Isolation and Detection of Extracellular Vesicles for Cancer Diagnostics. Adv. Healthc. Mater. 2018, 7, e1800484. [Google Scholar] [CrossRef]
- Iwai, K.; Minamisawa, T.; Suga, K.; Yajima, Y.; Shiba, K. Isolation of human salivary extracellular vesicles by iodixanol density gradient ultracentrifugation and their characterizations. J. Extracell. Vesicles 2016, 5, 30829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, P.G.; Lee, J.E.; You, S.; Kim, T.K.; Cho, J.H.; Kim, I.S.; Kwon, T.H.; Kim, C.D.; Park, S.H.; Hwang, D.; et al. Proteomic analysis of urinary exosomes from patients of early IgA nephropathy and thin basement membrane nephropathy. Proteomics 2011, 11, 2459–2475. [Google Scholar] [CrossRef] [PubMed]
- Musante, L.; Saraswat, M.; Duriez, E.; Byrne, B.; Ravida, A.; Domon, B.; Holthofer, H. Biochemical and physical characterisation of urinary nanovesicles following CHAPS treatment. PLoS ONE 2012, 7, e37279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hildonen, S.; Skarpen, E.; Halvorsen, T.G.; Reubsaet, L. Isolation and mass spectrometry analysis of urinary extraexosomal proteins. Sci. Rep. 2016, 6, 36331. [Google Scholar] [CrossRef] [PubMed]
- Barrachina, M.N.; Sueiro, A.M.; Casas, V.; Izquierdo, I.; Hermida-Nogueira, L.; Guitian, E.; Casanueva, F.F.; Abian, J.; Carrascal, M.; Pardo, M.; et al. A Combination of Proteomic Approaches Identifies A Panel of Circulating Extracellular Vesicle Proteins Related to the Risk of Suffering Cardiovascular Disease in Obese Patients. Proteomics 2019, 19, e1800248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merchant, M.L.; Powell, D.W.; Wilkey, D.W.; Cummins, T.D.; Deegens, J.K.; Rood, I.M.; McAfee, K.J.; Fleischer, C.; Klein, E.; Klein, J.B. Microfiltration isolation of human urinary exosomes for characterization by MS. Proteom. Clin. Appl. 2010, 4, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, M.L.; Ilmer, M.; Silva, L.P.; Hawke, D.H.; Recio, A.; Vorontsova, M.A.; Alt, E.; Vykoukal, J. Benchtop isolation and characterization of functional exosomes by sequential filtration. J. Chromatogr. A 2014, 1371, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Busatto, S.; Vilanilam, G.; Ticer, T.; Lin, W.L.; Dickson, D.W.; Shapiro, S.; Bergese, P.; Wolfram, J. Tangential Flow Filtration for Highly Efficient Concentration of Extracellular Vesicles from Large Volumes of Fluid. Cells 2018, 7, 273. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.S.; Lee, J.C.; Byeon, S.K.; Rha, K.H.; Moon, M.H. Size Dependent Lipidomic Analysis of Urinary Exosomes from Patients with Prostate Cancer by Flow Field-Flow Fractionation and Nanoflow Liquid Chromatography-Tandem Mass Spectrometry. Anal. Chem. 2017, 89, 2488–2496. [Google Scholar] [CrossRef]
- Hu, S.; Musante, L.; Tataruch, D.; Xu, X.; Kretz, O.; Henry, M.; Meleady, P.; Luo, H.; Zou, H.; Jiang, Y.; et al. Purification and Identification of Membrane Proteins from Urinary Extracellular Vesicles using Triton X-114 Phase Partitioning. J. Proteome Res. 2018, 17, 86–96. [Google Scholar] [CrossRef]
- Osti, D.; Del Bene, M.; Rappa, G.; Santos, M.; Matafora, V.; Richichi, C.; Faletti, S.; Beznoussenko, G.V.; Mironov, A.; Bachi, A.; et al. Clinical Significance of Extracellular Vesicles in Plasma from Glioblastoma Patients. Clin. Cancer Res. 2019, 25, 266–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smolarz, M.; Pietrowska, M.; Matysiak, N.; Mielanczyk, L.; Widlak, P. Proteome Profiling of Exosomes Purified from a Small Amount of Human Serum: The Problem of Co-Purified Serum Components. Proteomes 2019, 7, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navajas, R.; Corrales, F.J.; Paradela, A. Serum Exosome Isolation by Size-Exclusion Chromatography for the Discovery and Validation of Preeclampsia-Associated Biomarkers. Methods Mol. Biol. 2019, 1959, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, L.; Ludwig, S.; Schuler, P.J.; Hoffmann, T.K.; Brunner, C.; Theodoraki, M.N. The Potential of CD16 on Plasma-Derived Exosomes as a Liquid Biomarker in Head and Neck Cancer. Int. J. Mol. Sci. 2020, 21, 3739. [Google Scholar] [CrossRef]
- Ueda, K.; Ishikawa, N.; Tatsuguchi, A.; Saichi, N.; Fujii, R.; Nakagawa, H. Antibody-coupled monolithic silica microtips for highthroughput molecular profiling of circulating exosomes. Sci. Rep. 2014, 4, 6232. [Google Scholar] [CrossRef]
- Lim, J.; Choi, M.; Lee, H.; Kim, Y.H.; Han, J.Y.; Lee, E.S.; Cho, Y. Direct isolation and characterization of circulating exosomes from biological samples using magnetic nanowires. J. Nanobiotechnol. 2019, 17, 1. [Google Scholar] [CrossRef]
- Tauro, B.J.; Greening, D.W.; Mathias, R.A.; Mathivanan, S.; Ji, H.; Simpson, R.J. Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids. Mol. Cell Proteom. 2013, 12, 587–598. [Google Scholar] [CrossRef] [Green Version]
- Rupp, A.K.; Rupp, C.; Keller, S.; Brase, J.C.; Ehehalt, R.; Fogel, M.; Moldenhauer, G.; Marme, F.; Sultmann, H.; Altevogt, P. Loss of EpCAM expression in breast cancer derived serum exosomes: Role of proteolytic cleavage. Gynecol. Oncol. 2011, 122, 437–446. [Google Scholar] [CrossRef]
- Ghosh, A.; Davey, M.; Chute, I.C.; Griffiths, S.G.; Lewis, S.; Chacko, S.; Barnett, D.; Crapoulet, N.; Fournier, S.; Joy, A.; et al. Rapid isolation of extracellular vesicles from cell culture and biological fluids using a synthetic peptide with specific affinity for heat shock proteins. PLoS ONE 2014, 9, e110443. [Google Scholar] [CrossRef] [Green Version]
- Knol, J.C.; de Reus, I.; Schelfhorst, T.; Beekhof, R.; de Wit, M.; Piersma, S.R.; Pham, T.V.; Smit, E.F.; Verheul, H.M.W.; Jimenez, C.R. Peptide-mediated ‘miniprep’ isolation of extracellular vesicles is suitable for high-throughput proteomics. EuPA Open Proteom. 2016, 11, 11–15. [Google Scholar] [CrossRef] [Green Version]
- Balaj, L.; Atai, N.A.; Chen, W.; Mu, D.; Tannous, B.A.; Breakefield, X.O.; Skog, J.; Maguire, C.A. Heparin affinity purification of extracellular vesicles. Sci. Rep. 2015, 5, 10266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, F.; Jiao, F.; Xia, C.; Zhao, Y.; Ying, W.; Xie, Y.; Guan, X.; Tao, M.; Zhang, Y.; Qin, W.; et al. A novel strategy for facile serum exosome isolation based on specific interactions between phospholipid bilayers and TiO2. Chem. Sci. 2019, 10, 1579–1588. [Google Scholar] [CrossRef] [Green Version]
- Nakai, W.; Yoshida, T.; Diez, D.; Miyatake, Y.; Nishibu, T.; Imawaka, N.; Naruse, K.; Sadamura, Y.; Hanayama, R. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci. Rep. 2016, 6, 33935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, P.N.; Yin, H. Polymer-Based Purification of Extracellular Vesicles. Methods Mol. Biol. 2017, 1660, 91–103. [Google Scholar] [CrossRef]
- Weng, Y.; Sui, Z.; Shan, Y.; Hu, Y.; Chen, Y.; Zhang, L.; Zhang, Y. Effective isolation of exosomes with polyethylene glycol from cell culture supernatant for in-depth proteome profiling. Analyst 2016, 141, 4640–4646. [Google Scholar] [CrossRef] [PubMed]
- Macias, M.; Rebmann, V.; Mateos, B.; Varo, N.; Perez-Gracia, J.L.; Alegre, E.; Gonzalez, A. Comparison of six commercial serum exosome isolation methods suitable for clinical laboratories. Effect in cytokine analysis. Clin. Chem. Lab Med. 2019, 57, 1539–1545. [Google Scholar] [CrossRef]
- Taylor, D.D.; Zacharias, W.; Gercel-Taylor, C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol. Biol. 2011, 728, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Deregibus, M.C.; Figliolini, F.; D’Antico, S.; Manzini, P.M.; Pasquino, C.; De Lena, M.; Tetta, C.; Brizzi, M.F.; Camussi, G. Charge-based precipitation of extracellular vesicles. Int. J. Mol. Med. 2016, 38, 1359–1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, D.D.; Shah, S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods 2015, 87, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Geis-Asteggiante, L.; Dhabaria, A.; Edwards, N.; Ostrand-Rosenberg, S.; Fenselau, C. Top-down analysis of low mass proteins in exosomes shed by murine myeloid-derived suppressor cells. Int. J. Mass Spectrom. 2015, 378, 264–269. [Google Scholar] [CrossRef] [Green Version]
- Barrachina, M.N.; Calderon-Cruz, B.; Fernandez-Rocca, L.; Garcia, A. Application of Extracellular Vesicles Proteomics to Cardiovascular Disease: Guidelines, Data Analysis, and Future Perspectives. Proteomics 2019, 19, e1800247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joy, A.P.; Ayre, D.C.; Chute, I.C.; Beauregard, A.P.; Wajnberg, G.; Ghosh, A.; Lewis, S.M.; Ouellette, R.J.; Barnett, D.A. Proteome profiling of extracellular vesicles captured with the affinity peptide Vn96: Comparison of Laemmli and TRIzol(c) protein-extraction methods. J. Extracell. Vesicles 2018, 7, 1438727. [Google Scholar] [CrossRef] [PubMed]
- Kasprzyk, J.; Stepien, E.; Piekoszewski, W. Application of nano-LC-MALDI-TOF/TOF-MS for proteomic analysis of microvesicles. Clin. Biochem. 2017, 50, 241–243. [Google Scholar] [CrossRef]
- Fel, A.; Lewandowska, A.E.; Petrides, P.E.; Wisniewski, J.R. Comparison of Proteome Composition of Serum Enriched in Extracellular Vesicles Isolated from Polycythemia Vera Patients and Healthy Controls. Proteomes 2019, 7, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, X.F.; Chu, H.J.; Xu, Y.F.; Hua, L.; Wang, Z.P.; Huang, P.; Jia, H.L.; Zhang, L. Proteomics study of serum exosomes in Kawasaki disease patients with coronary artery aneurysms. Cardiol. J. 2019, 26, 584–593. [Google Scholar] [CrossRef] [Green Version]
- Shiromizu, T.; Kume, H.; Ishida, M.; Adachi, J.; Kano, M.; Matsubara, H.; Tomonaga, T. Quantitation of putative colorectal cancer biomarker candidates in serum extracellular vesicles by targeted proteomics. Sci. Rep. 2017, 7, 12782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kittivorapart, J.; Crew, V.K.; Wilson, M.C.; Heesom, K.J.; Siritanaratkul, N.; Toye, A.M. Quantitative proteomics of plasma vesicles identify novel biomarkers for hemoglobin E/beta-thalassemic patients. Blood Adv. 2018, 2, 95–104. [Google Scholar] [CrossRef] [Green Version]
- Sok Hwee Cheow, E.; Hwan Sim, K.; de Kleijn, D.; Neng Lee, C.; Sorokin, V.; Sze, S.K. Simultaneous Enrichment of Plasma Soluble and Extracellular Vesicular Glycoproteins Using Prolonged Ultracentrifugation-Electrostatic Repulsion-hydrophilic Interaction Chromatography (PUC-ERLIC) Approach. Mol. Cell Proteom. 2015, 14, 1657–1671. [Google Scholar] [CrossRef] [Green Version]
- Chen, I.H.; Xue, L.; Hsu, C.C.; Paez, J.S.; Pan, L.; Andaluz, H.; Wendt, M.K.; Iliuk, A.B.; Zhu, J.K.; Tao, W.A. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 3175–3180. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Li, L.; Iliuk, A.; Tao, W.A. Highly Efficient Phosphoproteome Capture and Analysis from Urinary Extracellular Vesicles. J. Proteome Res. 2018, 17, 3308–3316. [Google Scholar] [CrossRef]
- Jung, H.Y.; Lee, C.H.; Choi, J.Y.; Cho, J.H.; Park, S.H.; Kim, Y.L.; Moon, P.G.; Baek, M.C.; Berm Park, J.; Hoon Kim, Y.; et al. Potential urinary extracellular vesicle protein biomarkers of chronic active antibody-mediated rejection in kidney transplant recipients. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2020, 1138, 121958. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Pinna, R.; Gonzalez de Peredo, A.; Monsarrat, B.; Burlet-Schiltz, O.; Martin-Ventura, J.L. Label-free quantitative proteomic analysis of human plasma-derived microvesicles to find protein signatures of abdominal aortic aneurysms. Proteom. Clin. Appl. 2014, 8, 620–625. [Google Scholar] [CrossRef]
- Braga-Lagache, S.; Buchs, N.; Iacovache, M.I.; Zuber, B.; Jackson, C.B.; Heller, M. Robust Label-free, Quantitative Profiling of Circulating Plasma Microparticle (MP) Associated Proteins. Mol. Cell Proteom. 2016, 15, 3640–3652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harshman, S.W.; Canella, A.; Ciarlariello, P.D.; Agarwal, K.; Branson, O.E.; Rocci, A.; Cordero, H.; Phelps, M.A.; Hade, E.M.; Dubovsky, J.A.; et al. Proteomic characterization of circulating extracellular vesicles identifies novel serum myeloma associated markers. J. Proteom. 2016, 136, 89–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramacciotti, E.; Hawley, A.E.; Wrobleski, S.K.; Myers, D.D., Jr.; Strahler, J.R.; Andrews, P.C.; Guire, K.E.; Henke, P.K.; Wakefield, T.W. Proteomics of microparticles after deep venous thrombosis. Thromb. Res. 2010, 125, e269–e274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; Xu, K.; Zhou, B.; Chen, T.; Huang, Y.; Li, Q.; Wen, F.; Ge, W.; Wang, J.; Yu, S.; et al. A circulating extracellular vesicles-based novel screening tool for colorectal cancer revealed by shotgun and data-independent acquisition mass spectrometry. J. Extracell. Vesicles 2020, 9, 1750202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, K.; Kume, H.; Matsuzaki, K.; Kawashima, A.; Ujike, T.; Nagahara, A.; Uemura, M.; Miyagawa, Y.; Tomonaga, T.; Nonomura, N. Proteomic analysis of urinary extracellular vesicles from high Gleason score prostate cancer. Sci. Rep. 2017, 7, 42961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.; Liang, A.; He, Y.; Li, Z.; Li, Z.; Wang, G.; Sun, F. Proteomic analysis of seminal extracellular vesicle proteins involved in asthenozoospermia by iTRAQ. Mol. Reprod. Dev. 2019, 86, 1094–1105. [Google Scholar] [CrossRef]
- Fricke, F.; Michalak, M.; Warnken, U.; Hausser, I.; Schnolzer, M.; Kopitz, J.; Gebert, J. SILAC-Based Quantification of TGFBR2-Regulated Protein Expression in Extracellular Vesicles of Microsatellite Unstable Colorectal Cancers. Int. J. Mol. Sci. 2019, 20, 4162. [Google Scholar] [CrossRef] [Green Version]
- Harel, M.; Geiger, T. Plasma Biomarker Identification and Quantification by Microparticle Proteomics. Methods Mol. Biol. 2017, 1619, 477–486. [Google Scholar] [CrossRef]
- Wang, T.; Anderson, K.W.; Turko, I.V. Assessment of Extracellular Vesicles Purity Using Proteomic Standards. Anal. Chem. 2017, 89, 11070–11075. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Corbett, A.L.; Taatizadeh, E.; Tasnim, N.; Little, J.P.; Garnis, C.; Daugaard, M.; Guns, E.; Hoorfar, M.; Li, I.T.S. Challenges and opportunities in exosome research-Perspectives from biology, engineering, and cancer therapy. APL Bioeng. 2019, 3, 011503. [Google Scholar] [CrossRef] [Green Version]
- Bellin, G.; Gardin, C.; Ferroni, L.; Chachques, J.C.; Rogante, M.; Mitrecic, D.; Ferrari, R.; Zavan, B. Exosome in Cardiovascular Diseases: A Complex World Full of Hope. Cells 2019, 8, 166. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.; Xu, Y.; Ding, J.; Dong, J.; Jia, N.; Li, Y.; Zhang, M. Roles and Clinical Applications of Exosomes in Cardiovascular Disease. Biomed. Res. Int. 2020, 2020, 5424281. [Google Scholar] [CrossRef] [PubMed]
- Erozenci, L.A.; Bottger, F.; Bijnsdorp, I.V.; Jimenez, C.R. Urinary exosomal proteins as (pan-)cancer biomarkers: Insights from the proteome. FEBS Lett. 2019, 593, 1580–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chettimada, S.; Lorenz, D.R.; Misra, V.; Dillon, S.T.; Reeves, R.K.; Manickam, C.; Morgello, S.; Kirk, G.D.; Mehta, S.H.; Gabuzda, D. Exosome markers associated with immune activation and oxidative stress in HIV patients on antiretroviral therapy. Sci. Rep. 2018, 8, 7227. [Google Scholar] [CrossRef] [PubMed]
- Vallabhajosyula, P.; Korutla, L.; Habertheuer, A.; Yu, M.; Rostami, S.; Yuan, C.X.; Reddy, S.; Liu, C.; Korutla, V.; Koeberlein, B.; et al. Tissue-specific exosome biomarkers for noninvasively monitoring immunologic rejection of transplanted tissue. J. Clin. Investig. 2017, 127, 1375–1391. [Google Scholar] [CrossRef] [Green Version]
- Mathivanan, S.; Lim, J.W.; Tauro, B.J.; Ji, H.; Moritz, R.L.; Simpson, R.J. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol. Cell Proteom. 2010, 9, 197–208. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.Q.; Lee, D.; Kim, Y.; Paek, M.; Kim, M.; Jang, K.S.; Oh, J.; Lee, Y.S.; Yeon, J.E.; Lubman, D.M.; et al. Platelet Factor 4 as a Novel Exosome Marker in MALDI-MS Analysis of Exosomes from Human Serum. Anal. Chem. 2019, 91, 13297–13305. [Google Scholar] [CrossRef]
- Waldenstrom, A.; Genneback, N.; Hellman, U.; Ronquist, G. Cardiomyocyte microvesicles contain DNA/RNA and convey biological messages to target cells. PLoS ONE 2012, 7, e34653. [Google Scholar] [CrossRef] [Green Version]
- Liang, B.; He, X.; Zhao, Y.X.; Zhang, X.X.; Gu, N. Advances in Exosomes Derived from Different Cell Sources and Cardiovascular Diseases. Biomed. Res. Int. 2020, 2020, 7298687. [Google Scholar] [CrossRef]
- Reddy, V.S.; Madala, S.K.; Trinath, J.; Reddy, G.B. Extracellular small heat shock proteins: Exosomal biogenesis and function. Cell Stress Chaperones 2018, 23, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Tang, H.; Mitchell-Silbaugh, K.; Fang, X.; Han, Z.; Ouyang, K. Heat Shock Protein 60 in Cardiovascular Physiology and Diseases. Front. Mol. Biosci. 2020, 7, 73. [Google Scholar] [CrossRef] [PubMed]
- Malik, Z.A.; Kott, K.S.; Poe, A.J.; Kuo, T.; Chen, L.; Ferrara, K.W.; Knowlton, A.A. Cardiac myocyte exosomes: Stability, HSP60, and proteomics. Am. J. Physiol. Heart Circ. Physiol. 2013, 304, H954–H965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsis, T.; Pierouli, K.; Diakou, K.L.; Papakonstantinou, E.; Bacopoulou, F.; Chrousos, G.P.; Vlachakis, D. Exosomics. EMBnet J. 2020, 26. [Google Scholar] [CrossRef]
- Gupta, S.; Knowlton, A.A. HSP60 trafficking in adult cardiac myocytes: Role of the exosomal pathway. Am. J. Physiol. Heart Circ. Physiol. 2007, 292, H3052–H3056. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Deng, L.; Wang, D.; Li, N.; Chen, X.; Cheng, X.; Yuan, J.; Gao, X.; Liao, M.; Wang, M.; et al. Mechanism of TNF-alpha autocrine effects in hypoxic cardiomyocytes: Initiated by hypoxia inducible factor 1alpha, presented by exosomes. J. Mol. Cell Cardiol. 2012, 53, 848–857. [Google Scholar] [CrossRef]
- Cheow, E.S.; Cheng, W.C.; Lee, C.N.; de Kleijn, D.; Sorokin, V.; Sze, S.K. Plasma-derived Extracellular Vesicles Contain Predictive Biomarkers and Potential Therapeutic Targets for Myocardial Ischemic (MI) Injury. Mol. Cell Proteom. 2016, 15, 2628–2640. [Google Scholar] [CrossRef] [Green Version]
- Foglio, E.; Puddighinu, G.; Fasanaro, P.; D’Arcangelo, D.; Perrone, G.A.; Mocini, D.; Campanella, C.; Coppola, L.; Logozzi, M.; Azzarito, T.; et al. Exosomal clusterin, identified in the pericardial fluid, improves myocardial performance following MI through epicardial activation, enhanced arteriogenesis and reduced apoptosis. Int. J. Cardiol. 2015, 197, 333–347. [Google Scholar] [CrossRef]
- Yang, N.; Qin, Q. Apolipoprotein J: A New Predictor and Therapeutic Target in Cardiovascular Disease? Chin. Med. J. 2015, 128, 2530–2534. [Google Scholar] [CrossRef]
- Martinez-Bujidos, M.; Rull, A.; Gonzalez-Cura, B.; Perez-Cuellar, M.; Montoliu-Gaya, L.; Villegas, S.; Ordonez-Llanos, J.; Sanchez-Quesada, J.L. Clusterin/apolipoprotein J binds to aggregated LDL in human plasma and plays a protective role against LDL aggregation. FASEB J. 2015, 29, 1688–1700. [Google Scholar] [CrossRef]
- Vicencio, J.M.; Yellon, D.M.; Sivaraman, V.; Das, D.; Boi-Doku, C.; Arjun, S.; Zheng, Y.; Riquelme, J.A.; Kearney, J.; Sharma, V.; et al. Plasma exosomes protect the myocardium from ischemia-reperfusion injury. J. Am. Coll. Cardiol. 2015, 65, 1525–1536. [Google Scholar] [CrossRef] [Green Version]
- Elakabawi, K.; Lin, J.; Jiao, F.; Guo, N.; Yuan, Z. Kawasaki Disease: Global Burden and Genetic Background. Cardiol. Res. 2020, 11, 9–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Song, Q.F.; Jin, J.J.; Huang, P.; Wang, Z.P.; Xie, X.F.; Gu, X.Q.; Gao, X.J.; Jia, H.L. Differential protein analysis of serum exosomes post-intravenous immunoglobulin therapy in patients with Kawasaki disease. Cardiol. Young 2017, 27, 1786–1796. [Google Scholar] [CrossRef] [PubMed]
- Kennel, P.J.; Saha, A.; Maldonado, D.A.; Givens, R.; Brunjes, D.L.; Castillero, E.; Zhang, X.; Ji, R.; Yahi, A.; George, I.; et al. Serum exosomal protein profiling for the non-invasive detection of cardiac allograft rejection. J. Heart Lung Transpl. 2018, 37, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Lai, R.C.; Arslan, F.; Lee, M.M.; Sze, N.S.; Choo, A.; Chen, T.S.; Salto-Tellez, M.; Timmers, L.; Lee, C.N.; El Oakley, R.M.; et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010, 4, 214–222. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Zhao, Y.; Sun, L.; Sun, X.; Zhao, X.; Sun, X.; Qian, H.; Xu, W.; Zhu, W. Exosomes Derived from Akt-Modified Human Umbilical Cord Mesenchymal Stem Cells Improve Cardiac Regeneration and Promote Angiogenesis via Activating Platelet-Derived Growth Factor D. Stem Cells Transl. Med. 2017, 6, 51–59. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Jin, X.; Hu, C.F.; Li, R.; Zhou, Z.; Shen, C.X. Exosomes Derived from Mesenchymal Stem Cells Rescue Myocardial Ischaemia/Reperfusion Injury by Inducing Cardiomyocyte Autophagy Via AMPK and Akt Pathways. Cell Physiol. Biochem. 2017, 43, 52–68. [Google Scholar] [CrossRef]
- Anderson, J.D.; Johansson, H.J.; Graham, C.S.; Vesterlund, M.; Pham, M.T.; Bramlett, C.S.; Montgomery, E.N.; Mellema, M.S.; Bardini, R.L.; Contreras, Z.; et al. Comprehensive Proteomic Analysis of Mesenchymal Stem Cell Exosomes Reveals Modulation of Angiogenesis via Nuclear Factor-KappaB Signaling. Stem Cells 2016, 34, 601–613. [Google Scholar] [CrossRef] [Green Version]
- Cosme, J.; Guo, H.; Hadipour-Lakmehsari, S.; Emili, A.; Gramolini, A.O. Hypoxia-Induced Changes in the Fibroblast Secretome, Exosome, and Whole-Cell Proteome Using Cultured, Cardiac-Derived Cells Isolated from Neonatal Mice. J. Proteome Res. 2017, 16, 2836–2847. [Google Scholar] [CrossRef]
- Lyu, L.; Wang, H.; Li, B.; Qin, Q.; Qi, L.; Nagarkatti, M.; Nagarkatti, P.; Janicki, J.S.; Wang, X.L.; Cui, T. A critical role of cardiac fibroblast-derived exosomes in activating renin angiotensin system in cardiomyocytes. J. Mol. Cell Cardiol. 2015, 89, 268–279. [Google Scholar] [CrossRef] [Green Version]
- Sheldon, H.; Heikamp, E.; Turley, H.; Dragovic, R.; Thomas, P.; Oon, C.E.; Leek, R.; Edelmann, M.; Kessler, B.; Sainson, R.C.; et al. New mechanism for Notch signaling to endothelium at a distance by Delta-like 4 incorporation into exosomes. Blood 2010, 116, 2385–2394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, H.; Shi, S.; Wang, S.; Peng, H.; Ding, S.J.; Wang, L. Proteomic Profiling Exosomes from Vascular Smooth Muscle Cell. Proteom. Clin. Appl. 2018, 12, e1700097. [Google Scholar] [CrossRef] [PubMed]
- Kapustin, A.N.; Chatrou, M.L.; Drozdov, I.; Zheng, Y.; Davidson, S.M.; Soong, D.; Furmanik, M.; Sanchis, P.; De Rosales, R.T.; Alvarez-Hernandez, D.; et al. Vascular smooth muscle cell calcification is mediated by regulated exosome secretion. Circ. Res. 2015, 116, 1312–1323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapustin, A.N.; Schoppet, M.; Schurgers, L.J.; Reynolds, J.L.; McNair, R.; Heiss, A.; Jahnen-Dechent, W.; Hackeng, T.M.; Schlieper, G.; Harrison, P.; et al. Prothrombin Loading of Vascular Smooth Muscle Cell-Derived Exosomes Regulates Coagulation and Calcification. Arterioscler. Thromb. Vasc. Biol. 2017, 37, e22–e32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barile, L.; Cervio, E.; Lionetti, V.; Milano, G.; Ciullo, A.; Biemmi, V.; Bolis, S.; Altomare, C.; Matteucci, M.; Di Silvestre, D.; et al. Cardioprotection by cardiac progenitor cell-secreted exosomes: Role of pregnancy-associated plasma protein-A. Cardiovasc. Res. 2018, 114, 992–1005. [Google Scholar] [CrossRef] [Green Version]



| EV Subclasses | ||||
|---|---|---|---|---|
| EVs * | Exosomes | Microvesicles | Apoptotic Bodies | |
| DIAMETER | Small EVs: <100 or <200 nm; Medium or large EVs: >200 nm | 30–150 nm | 50–1000 nm and more | 500–4000 nm |
| FLOATING DENSITY | Low density; Middle density; High density | Overlap in exosome and microvesicle density (1.08–1.19 g/mL) makes it difficult to distinguish the two groups [8] | 1.16–1.28 g/mL [9] | |
| BIOGENESIS | Naturally released from the cells. The EV assignment to a particular biogenesis pathway is sometimes difficult unless the use of live imaging techniques. | Multivesicular endosome | Plasma membrane budding and cleavage | Blebs detaching from dying apoptotic cell |
| MARKERS |
| ESCRT complex, tetraspanins (CD63, CD9, CD81, and CD82), flotillins, TSG101, ALIX, heat shock proteins (HSC70, HSP60, HSP70, HSPA5, CCT2, and HSP90) | Annexin A1, annexin V, flotillin-2, selectins, integrins, CD40 | Phosphatidylserine, histones, annexin V, TSP, and C3b |
| Isolation Techniques | Advantages | Limitations |
|---|---|---|
| Differential ultracentrifugation |
|
|
| Density-Gradient Ultracentrifugation |
|
|
| Size-based isolation |
|
|
| Immunoaffinity isolation |
|
|
| Polymer-induced precipitation |
|
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mallia, A.; Gianazza, E.; Zoanni, B.; Brioschi, M.; Barbieri, S.S.; Banfi, C. Proteomics of Extracellular Vesicles: Update on Their Composition, Biological Roles and Potential Use as Diagnostic Tools in Atherosclerotic Cardiovascular Diseases. Diagnostics 2020, 10, 843. https://doi.org/10.3390/diagnostics10100843
Mallia A, Gianazza E, Zoanni B, Brioschi M, Barbieri SS, Banfi C. Proteomics of Extracellular Vesicles: Update on Their Composition, Biological Roles and Potential Use as Diagnostic Tools in Atherosclerotic Cardiovascular Diseases. Diagnostics. 2020; 10(10):843. https://doi.org/10.3390/diagnostics10100843
Chicago/Turabian StyleMallia, Alice, Erica Gianazza, Beatrice Zoanni, Maura Brioschi, Silvia Stella Barbieri, and Cristina Banfi. 2020. "Proteomics of Extracellular Vesicles: Update on Their Composition, Biological Roles and Potential Use as Diagnostic Tools in Atherosclerotic Cardiovascular Diseases" Diagnostics 10, no. 10: 843. https://doi.org/10.3390/diagnostics10100843