Dysregulated microRNA Expression Relevant to TERT Promoter Mutations in Tonsil Cancer—A Pilot Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients, Samples, and Clinicopathological and Genetic Mutational Data
2.2. MicroRNA Extraction and Nanostring nCounter miRNA Expression Assay
2.3. Function Enrichment Analysis
2.4. Statistical Analyses
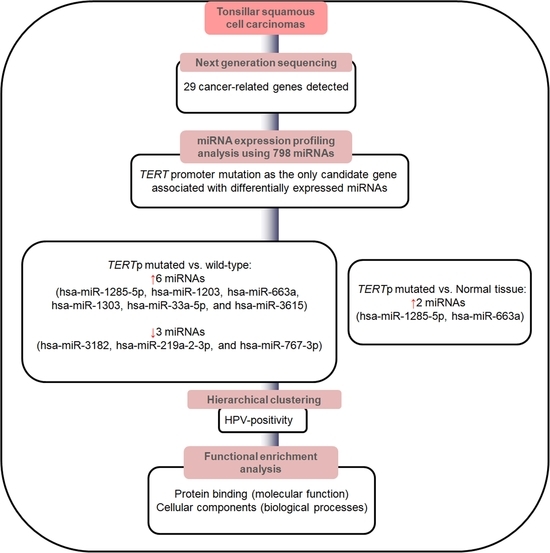
3. Results
3.1. Baseline Characteristics
3.2. Screening of Candidate miRNAs Associated with Specific Genetic Mutations
3.3. Identification of Potential Functional Pathways Related to miRNAs
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Taberna, M.; Mena, M.; Pavon, M.A.; Alemany, L.; Gillison, M.L.; Mesia, R. Human papillomavirus-related oropharyngeal cancer. Ann. Oncol. 2017, 28, 2386–2398. [Google Scholar] [CrossRef]
- Luginbuhl, A.; Sanders, M.; Spiro, J.D. Prevalence, morphology, and prognosis of human papillomavirus in tonsillar cancer. Ann. Otol. Rhinol. Laryngol. 2009, 118, 742–749. [Google Scholar] [CrossRef]
- Olaleye, O.; Moorthy, R.; Lyne, O.; Black, M.; Mitchell, D.; Wiseberg, J. A 20-year retrospective study of tonsil cancer incidence and survival trends in South East England: 1987–2006. Clin. Otolaryngol. 2011, 36, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Throat Cancer: Squamous Cell Carcinoma of the Tonsil. Available online: https://healthengine.com.au/info/throat-cancer-squamous-cell-carcinoma-of-the-tonsil (accessed on 25 January 2023).
- Kuo, Y.Y.; Chu, P.Y.; Chang, S.Y.; Wang, Y.F.; Tsai, T.L.; Yang, M.H.; Wang, L.W.; Tai, S.K. Treatment selection for tonsillar squamous cell carcinoma. J. Chin. Med. Assoc. 2013, 76, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.B.; Edge, S.B.; Greene, F.L.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R.; Sullivan, D.C. AJCC Cancer Staging Manual; Springer International Publishing: Cham, Switzerland, 2017; Volume 1024. [Google Scholar]
- Timbang, M.R.; Sim, M.W.; Bewley, A.F.; Farwell, D.G.; Mantravadi, A.; Moore, M.G. HPV-related oropharyngeal cancer: A review on burden of the disease and opportunities for prevention and early detection. Hum. Vaccines Immunother. 2019, 15, 1920–1928. [Google Scholar] [CrossRef] [PubMed]
- Doweck, I.; Robbins, K.T.; Mendenhall, W.M.; Hinerman, R.W.; Morris, C.; Amdur, R. Neck level-specific nodal metastases in oropharyngeal cancer: Is there a role for selective neck dissection after definitive radiation therapy? Head Neck 2003, 25, 960–967. [Google Scholar] [CrossRef]
- Slootweg, P.J.; Eveson, J.W. Tumours of the oral cavity and oropharynx. In World Health Organization Classification of Tumours: Head and Neck Tumours; Barnes, L., Eveson, J.W., Reichart, P., Sidransky, D., Eds.; International Agency for Research on Cancer (IARC): Lyon, France, 2005; pp. 163–207. [Google Scholar]
- Park, H.Y.; Lee, J.S.; Wee, J.H.; Kang, J.W.; Kim, E.S.; Koo, T.; Hwang, H.S.; Kim, H.J.; Kang, H.S.; Lim, H.; et al. Assessment of the Mutation Profile of Tonsillar Squamous Cell Carcinomas Using Targeted Next-Generation Sequencing. Biomedicines 2023, 11, 851. [Google Scholar] [CrossRef]
- Masuda, M.; Wakasaki, T.; Toh, S. Stress-triggered atavistic reprogramming (STAR) addiction: Driving force behind head and neck cancer? Am. J. Cancer Res. 2016, 6, 1149–1166. [Google Scholar]
- Dogan, S.; Xu, B.; Middha, S.; Vanderbilt, C.M.; Bowman, A.S.; Migliacci, J.; Morris, L.G.; Seshan, V.E.; Ganly, I. Identification of prognostic molecular biomarkers in 157 HPV-positive and HPV-negative squamous cell carcinomas of the oropharynx. Int. J. Cancer 2019, 145, 3152–3162. [Google Scholar] [CrossRef]
- Kim, H.; Kwon, M.J.; Park, B.; Choi, H.G.; Nam, E.S.; Cho, S.J.; Min, K.W.; Kim, E.S.; Hwang, H.S.; Hong, M.; et al. Negative Prognostic Implication of TERT Promoter Mutations in Human Papillomavirus-Negative Tonsillar Squamous Cell Carcinoma Under the New 8th AJCC Staging System. Indian J. Surg. Oncol. 2021, 12, 134–143. [Google Scholar] [CrossRef]
- Ying, S.Y.; Chang, D.C.; Lin, S.L. The microRNA (miRNA): Overview of the RNA genes that modulate gene function. Mol. Biotechnol. 2008, 38, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Lajer, C.B.; Nielsen, F.C.; Friis-Hansen, L.; Norrild, B.; Borup, R.; Garnaes, E.; Rossing, M.; Specht, L.; Therkildsen, M.H.; Nauntofte, B.; et al. Different miRNA signatures of oral and pharyngeal squamous cell carcinomas: A prospective translational study. Br. J. Cancer 2011, 104, 830–840. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gee, H.; Rose, B.; Lee, C.S.; Clark, J.; Elliott, M.; Gamble, J.R.; Cairns, M.J.; Harris, A.; Khoury, S.; et al. Regulation of the tumour suppressor PDCD4 by miR-499 and miR-21 in oropharyngeal cancers. BMC Cancer 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Vojtechova, Z.; Sabol, I.; Salakova, M.; Smahelova, J.; Zavadil, J.; Turek, L.; Grega, M.; Klozar, J.; Prochazka, B.; Tachezy, R. Comparison of the miRNA profiles in HPV-positive and HPV-negative tonsillar tumors and a model system of human keratinocyte clones. BMC Cancer 2016, 16, 382. [Google Scholar] [CrossRef]
- Vojtechova, Z.; Zavadil, J.; Klozar, J.; Grega, M.; Tachezy, R. Comparison of the miRNA expression profiles in fresh frozen and formalin-fixed paraffin-embedded tonsillar tumors. PLoS ONE 2017, 12, e0179645. [Google Scholar] [CrossRef]
- Quabius, E.S.; Merz, I.; Gorogh, T.; Hedderich, J.; Haag, J.; Rocken, C.; Ambrosch, P.; Hoffmann, M. miRNA-expression in tonsillar squamous cell carcinomas in relation to HPV infection and expression of the antileukoproteinase SLPI. Papillomavirus Res. 2017, 4, 26–34. [Google Scholar] [CrossRef]
- Bersani, C.; Mints, M.; Tertipis, N.; Haeggblom, L.; Nasman, A.; Romanitan, M.; Dalianis, T.; Ramqvist, T. MicroRNA-155, -185 and -193b as biomarkers in human papillomavirus positive and negative tonsillar and base of tongue squamous cell carcinoma. Oral Oncol. 2018, 82, 8–16. [Google Scholar] [CrossRef]
- Sais, D.; Zhang, X.; Marques, T.M.; Rose, B.; Khoury, S.; Hill, M.; Deutsch, F.; Lyons, J.G.; Gama-Carvalho, M.; Tran, N. Human papillomavirus 16 E6 modulates the expression of miR-496 in oropharyngeal cancer. Virology 2018, 521, 149–157. [Google Scholar] [CrossRef]
- Ramirez-Moya, J.; Wert-Lamas, L.; Riesco-Eizaguirre, G.; Santisteban, P. Impaired microRNA processing by DICER1 downregulation endows thyroid cancer with increased aggressiveness. Oncogene 2019, 38, 5486–5499. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Wixon, J.; Kell, D. The Kyoto encyclopedia of genes and genomes—KEGG. Yeast 2000, 17, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B Stat. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kim, H.S.; Kwon, M.J.; Song, J.H.; Kim, E.S.; Kim, H.Y.; Min, K.W. Clinical implications of TERT promoter mutation on IDH mutation and MGMT promoter methylation in diffuse gliomas. Pathol. Res. Pract. 2018, 214, 881–888. [Google Scholar] [CrossRef]
- Aubert, G.; Lansdorp, P.M. Telomeres and aging. Physiol. Rev. 2008, 88, 557–579. [Google Scholar] [CrossRef] [PubMed]
- Arita, H.; Narita, Y.; Takami, H.; Fukushima, S.; Matsushita, Y.; Yoshida, A.; Miyakita, Y.; Ohno, M.; Shibui, S.; Ichimura, K. TERT promoter mutations rather than methylation are the main mechanism for TERT upregulation in adult gliomas. Acta Neuropathol. 2013, 126, 939–941. [Google Scholar] [CrossRef]
- Vinothkumar, V.; Arunkumar, G.; Revathidevi, S.; Arun, K.; Manikandan, M.; Rao, A.K.; Rajkumar, K.S.; Ajay, C.; Rajaraman, R.; Ramani, R.; et al. TERT promoter hot spot mutations are frequent in Indian cervical and oral squamous cell carcinomas. Tumor Biol. 2016, 37, 7907–7913. [Google Scholar] [CrossRef]
- Barczak, W.; Suchorska, W.M.; Sobecka, A.; Bednarowicz, K.; Machczynski, P.; Golusinski, P.; Rubis, B.; Masternak, M.M.; Golusinski, W. hTERT C250T promoter mutation and telomere length as a molecular markers of cancer progression in patients with head and neck cancer. Mol. Med. Rep. 2017, 16, 441–446. [Google Scholar] [CrossRef]
- Chang, K.P.; Wang, C.I.; Pickering, C.R.; Huang, Y.; Tsai, C.N.; Tsang, N.M.; Kao, H.K.; Cheng, M.H.; Myers, J.N. Prevalence of promoter mutations in the TERT gene in oral cavity squamous cell carcinoma. Head Neck 2017, 39, 1131–1137. [Google Scholar] [CrossRef]
- Yuan, P.; Cao, J.-l.; Abuduwufuer, A.; Wang, L.-M.; Yuan, X.-S.; Lv, W.; Hu, J. Clinical characteristics and prognostic significance of TERT promoter mutations in cancer: A cohort study and a meta-analysis. PLoS ONE 2016, 11, e0146803. [Google Scholar] [CrossRef]
- Nunvar, J.; Pagacova, L.; Vojtechova, Z.; Azevedo, N.T.D.; Smahelova, J.; Salakova, M.; Tachezy, R. Lack of Conserved miRNA Deregulation in HPV-Induced Squamous Cell Carcinomas. Biomolecules 2021, 11, 764. [Google Scholar] [CrossRef]
- Miller, D.L.; Davis, J.W.; Taylor, K.H.; Johnson, J.; Shi, Z.; Williams, R.; Atasoy, U.; Lewis, J.S., Jr.; Stack, M.S. Identification of a human papillomavirus-associated oncogenic miRNA panel in human oropharyngeal squamous cell carcinoma validated by bioinformatics analysis of the Cancer Genome Atlas. Am. J. Pathol. 2015, 185, 679–692. [Google Scholar] [CrossRef]
- Annunziata, C.; Pezzuto, F.; Greggi, S.; Ionna, F.; Losito, S.; Botti, G.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Distinct profiles of TERT promoter mutations and telomerase expression in head and neck cancer and cervical carcinoma. Int. J. Cancer 2018, 143, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- Boscolo-Rizzo, P.; Giunco, S.; Rampazzo, E.; Brutti, M.; Spinato, G.; Menegaldo, A.; Stellin, M.; Mantovani, M.; Bandolin, L.; Rossi, M.; et al. TERT promoter hotspot mutations and their relationship with TERT levels and telomere erosion in patients with head and neck squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2020, 146, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Kim, G.; Cho, H.; Kim, S.; Lee, D.; Park, S.; Park, K.H.; Lee, H. Diagnostic performance of HPV E6/E7, hTERT, and Ki67 mRNA RT-qPCR assays on formalin-fixed paraffin-embedded cervical tissue specimens from women with cervical cancer. Exp. Mol. Pathol. 2015, 98, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Roberts, J.; Dakic, A.; Zhang, Y.; Schlegel, R. HPV E7 contributes to the telomerase activity of immortalized and tumorigenic cells and augments E6-induced hTERT promoter function. Virology 2008, 375, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Dakic, A.; Chen, R.; Disbrow, G.L.; Zhang, Y.; Dai, Y.; Schlegel, R. Cell-restricted immortalization by human papillomavirus correlates with telomerase activation and engagement of the hTERT promoter by Myc. J. Virol. 2008, 82, 11568–11576. [Google Scholar] [CrossRef]
- Zhou, S.; Zhang, Z.; Zheng, P.; Zhao, W.; Han, N. MicroRNA-1285-5p influences the proliferation and metastasis of non-small-cell lung carcinoma cells via downregulating CDH1 and Smad4. Tumor Biol. 2017, 39, 1010428317705513. [Google Scholar] [CrossRef]
- Borrelli, N.; Denaro, M.; Ugolini, C.; Poma, A.M.; Miccoli, M.; Vitti, P.; Miccoli, P.; Basolo, F. miRNA expression profiling of ‘noninvasive follicular thyroid neoplasms with papillary-like nuclear features’ compared with adenomas and infiltrative follicular variants of papillary thyroid carcinomas. Mod. Pathol. 2017, 30, 39–51. [Google Scholar] [CrossRef]
- Yi, C.; Wang, Q.; Wang, L.; Huang, Y.; Li, L.; Liu, L.; Zhou, X.; Xie, G.; Kang, T.; Wang, H.; et al. MiR-663, a microRNA targeting p21WAF1/CIP1, promotes the proliferation and tumorigenesis of nasopharyngeal carcinoma. Oncogene 2012, 31, 4421–4433. [Google Scholar] [CrossRef]
- Zhi-Yong, L.; Guang-Ling, Z.; Mei-Mei, W.; Ya-Nan, X.; He-Qin, C. MicroRNA-663 targets TGFB1 and regulates lung cancer proliferation. Asian Pac. J. Cancer Prev. 2011, 12, 2819–2823. [Google Scholar]
- Kuroda, K.; Fukuda, T.; Krstic-Demonacos, M.; Demonacos, C.; Okumura, K.; Isogai, H.; Hayashi, M.; Saito, K.; Isogai, E. miR-663a regulates growth of colon cancer cells, after administration of antimicrobial peptides, by targeting CXCR4-p21 pathway. BMC Cancer 2017, 17, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hidaka, H.; Seki, N.; Yoshino, H.; Yamasaki, T.; Yamada, Y.; Nohata, N.; Fuse, M.; Nakagawa, M.; Enokida, H. Tumor suppressive microRNA-1285 regulates novel molecular targets: Aberrant expression and functional significance in renal cell carcinoma. Oncotarget 2012, 3, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Hu, H.; Zhou, Z.; Sun, L.; Peng, L.; Yu, L.; Sun, L.; Liu, J.; Yang, Z.; Ran, Y. Tumor-suppressive mir-663 gene induces mitotic catastrophe growth arrest in human gastric cancer cells. Oncol. Rep. 2010, 24, 105–112. [Google Scholar] [PubMed]
- Mazzarello, P.; Bentivoglio, M. The centenarian Golgi apparatus. Nature 1998, 392, 543–544. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Pan, X.; Li, Z.; Chen, P.; Quan, J.; Lin, C.; Lai, Y.; Xu, J.; Xu, W.; Guan, X.; et al. Oncogenic miR-663a is associated with cellular function and poor prognosis in renal cell carcinoma. Biomed. Pharmacother. 2018, 105, 1155–1163. [Google Scholar] [CrossRef]
- Foye, C.; Yan, I.K.; David, W.; Shukla, N.; Habboush, Y.; Chase, L.; Ryland, K.; Kesari, V.; Patel, T. Comparison of miRNA quantitation by Nanostring in serum and plasma samples. PLoS ONE 2017, 12, e0189165. [Google Scholar] [CrossRef]



| Characteristic | n = 22 (%) | HPV-Negative n = 5 (22.7%) | HPV-Positive n = 17 (77.3%) | p-Value |
|---|---|---|---|---|
| Sex | ||||
| Male | 22 (100) | 5 (100) | 17 (100) | |
| Female | 0 (0.0%) | |||
| Age, mean ± SD (years) | 55.22 ± 9.35 (range, 41–73) | |||
| ≤60 | 15 (68.2) | 3 (60.0) | 12 (70.6) | 0.655 |
| >60 | 7 (31.8) | 2 (40.0) | 5 (29.4) | |
| Smoking | ||||
| Light | 7 (31.8) | 1 (20.0) | 6 (35.3) | 0.519 |
| Heavy | 15 (68.2) | 4 (80.0) | 11 (64.7) | |
| Alcohol | ||||
| Light | 11 (50) | 1 (20.0) | 10 (58.8) | 0.127 |
| Heavy | 11 (50) | 4 (80.0) | 7 (41.2) | |
| Lymphovascular invasion | ||||
| negative | 5 (22.7) | 1 (20.0) | 4 (23.5) | |
| positive | 17 (77.3) | 4 (80.0) | 13 (76.5) | 1000 |
| pT category | ||||
| T1–T2 | 12 (54.5) | 4 (80.0) | 8 (47.1) | 0.193 |
| T3–T4 | 10 (45.5) | 1 (20.0) | 9 (52.9) | |
| pNodal status | ||||
| N0–1 | 13 (59.1) | 1 (20.0) | 12 (70.6) | 0.116 |
| N2–3 | 9 (40.9) | 4 (80.0) | 5 (29.4) | |
| AJCC stage | ||||
| I–II | 15 (68.2) | 1 (20.0) | 10 (58.8) | 0.311 |
| III–IV | 7 (31.8) | 4 (80.0) | 7 (41.2) | |
| 5-year overall survival | ||||
| Survival | 12 (54.5) | 0 (0.0) | 12 (70.6) | 0.010 * |
| Death | 10 (45.5) | 5 (100.0) | 5 (29.4) | |
| 5-year disease-free survival | ||||
| Yes | 11 (50.0) | 0 (0.0) | 11 (64.7) | 0.035 * |
| No | 11 (50.0) | 5 (100.0) | 6 (35.3) | |
| Mutated detected tumors | ||||
| No detected | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| Detected | 22 (100) | 5 (100.0) | 17(100.0) | |
| TP53 | ||||
| Wild type | 8 (36.4) | 0 (0.0) | 8 (47.1) | 0.054 |
| Mutated | 14 (63.6) | 5 (100.0) | 9 (52.9) | |
| PICK3A | ||||
| Wild type | 12 (54.5) | 2 (40.0) | 10 (58.8) | 0.457 |
| Mutated | 10 (45.5) | 3 (60.0) | 7 (41.2) | |
| PTEN | ||||
| Wild type | 16 (72.7) | 4 (80.0) | 12 (70.6) | 0.678 |
| Mutated | 6 (27.3) | 1 (20.0) | 5 (29.4) | |
| SMAD4 | ||||
| Wild type | 17 (77.3) | 4 (80.0) | 14 (82.4) | 0.905 |
| Mutated | 5 (22.7) | 1 (20.0) | 3 (17.6) | |
| EGRF | ||||
| Wild type | 18 (81.8) | 3 (60.0) | 14 (82.4) | 0.294 |
| Mutated | 4 (18.2) | 2 (40.0) | 3 (17.6) | |
| TERTp | ||||
| Wild type | 20 (90.9) | 4 (80.0) | 16 (94.1) | 0.334 |
| Mutated | 2 (9.1) | 1 (20.0) | 1 (5.9) |
| TERTp-Mutated vs. Wild Type | Expression in Mutated Tumors | TERTp-Mutated vs. Normal Control | Expression in Mutated Tumors | |||
|---|---|---|---|---|---|---|
| Fold Change | FDR * | Fold Change | DE Call * | |||
| hsa-miR-1285-5p | 2.53 | 0.00 | up | 3.13 | Yes | up |
| hsa-miR-1203 | 1.56 | 0.01 | up | 2.83 | No | up |
| hsa-miR-3182 | −2.06 | 0.01 | down | −1.35 | No | down |
| hsa-miR-663a | 2.02 | 0.01 | up | 7.31 | Yes | up |
| hsa-miR-219a-2-3p | −1.57 | 0.01 | down | −1.29 | No | down |
| hsa-miR-767-3p | −1.52 | 0.01 | down | −1.72 | No | down |
| hsa-miR-1303 | 1.63 | 0.02 | up | 1.72 | No | up |
| hsa-miR-33a-5p | 1.42 | 0.04 | up | −2.4 | No | down |
| hsa-miR-3615 | 1.31 | 0.04 | up | 2.7 | No | up |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwon, M.J.; Park, H.Y.; Lee, J.S.; Kim, E.S.; Kim, N.Y.; Nam, E.S.; Cho, S.J.; Kang, H.S. Dysregulated microRNA Expression Relevant to TERT Promoter Mutations in Tonsil Cancer—A Pilot Study. Life 2023, 13, 2090. https://doi.org/10.3390/life13102090
Kwon MJ, Park HY, Lee JS, Kim ES, Kim NY, Nam ES, Cho SJ, Kang HS. Dysregulated microRNA Expression Relevant to TERT Promoter Mutations in Tonsil Cancer—A Pilot Study. Life. 2023; 13(10):2090. https://doi.org/10.3390/life13102090
Chicago/Turabian StyleKwon, Mi Jung, Ha Young Park, Joong Seob Lee, Eun Soo Kim, Nan Young Kim, Eun Sook Nam, Seong Jin Cho, and Ho Suk Kang. 2023. "Dysregulated microRNA Expression Relevant to TERT Promoter Mutations in Tonsil Cancer—A Pilot Study" Life 13, no. 10: 2090. https://doi.org/10.3390/life13102090