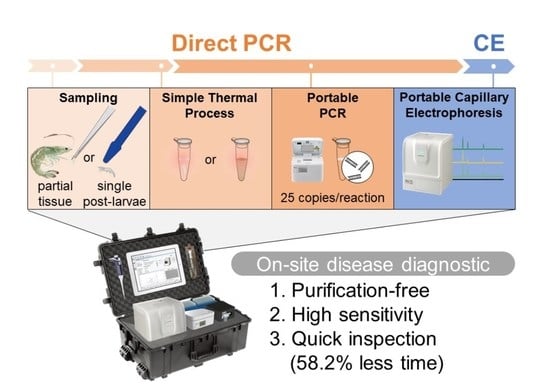
Combining Direct PCR Technology and Capillary Electrophoresis for an Easy-to-Operate and Highly Sensitive Infectious Disease Detection System for Shrimp
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. PCR kits and Equipments
2.3. Source and Tissue Sampling of Shrimp
2.4. Direct PCR for the Detection of Shrimp Disease
2.5. WSSV Detection in Juvenile Shrimp Using the IQ2000TM WSSV Detection System
2.6. Equations for Evaluating the Diagnostic Performance of Direct PCR-Based Capillary Electrophoresis
2.7. Ethics Statement
3. Results
3.1. Disease Detection in Shrimp Tissue Using Direct PCR
3.2. Increased Detection Limits Using Capillary Electrophoresis Analysis
3.3. Disease Detection in Adult Shrimp Tissue Using Direct PCR-Based Capillary Electrophoresis
3.4. Disease Detection in Individual Shrimp Post Larvae
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. A Quarterly Update on World Seafood Markets; FAO: Rome, Italy, 2019. [Google Scholar]
- Naylor, R.L.; Hardy, R.W.; Buschmann, A.H.; Bush, S.R.; Cao, L.; Klinger, D.H.; Little, D.C.; Lubchenco, J.; Shumway, S.E.; Troell, M. A 20-year retrospective review of global aquaculture. Nature 2021, 591, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, J.D.; Kazimierczak, J.; Sowińska, P.M.; Wójcik, E.A.; Siwicki, A.K.; Dastych, J. Growing trend of fighting infections in aquaculture environment−Opportunities and challenges of phage therapy. Antibiotics 2020, 9, 301. [Google Scholar] [CrossRef] [PubMed]
- Shinn, A.P.; Pratoomyot, J.; Griffiths, D.; Trong, T.Q.; Vu, N.T.; Jiravanichpaisal, P.; Briggs, M. Asian shrimp production and the economic costs of disease. Asian Fish. Sci. 2018, 31S, 29–58. [Google Scholar] [CrossRef]
- Tang, K.F.J.; Bondad-Reantaso, M.G.; Arthur, J.R.; MacKinnon, B.; Hao, B.; Alday-Sanz, V.; Liang, Y.; Dong, X. Shrimp Acute Hepatopancreatic Necrosis Disease Strategy Manual; No. 1190; FAO: Rome, Italy, 2020. [Google Scholar]
- Kumar, V.; Roy, S.; Behera, B.K.; Bossier, P.; Das, B.K. Acute hepatopancreatic necrosis disease (AHPND): Virulence, pathogenesis and mitigation strategies in shrimp aquaculture. Toxins 2021, 13, 524. [Google Scholar] [CrossRef] [PubMed]
- Oakey, J.; Smith, C.; Underwood, D.; Afsharnasab, M.; Alday-Sanz, V.; Dhar, A.; Sivakumar, S.; Sahul Hameed, A.S.; Beattie, K.; Crook, A. Global distribution of white spot syndrome virus genotypes determined using a novel genotyping assay. Arch. Virol. 2019, 164, 2061–2082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, L.Y.-H.; Yuan, B.; Convertino, M. COVID-19 non-pharmaceutical intervention portfolio effectiveness and risk communication predominance. Sci. Rep. 2021, 11, 10605. [Google Scholar] [CrossRef]
- Biao, X.; Yu, K. Shrimp farming in China: Operating characteristics, environmental impact and perspectives. Ocean Coast. Manag. 2007, 50, 538–550. [Google Scholar] [CrossRef]
- Iber, B.T.; Kasan, N.A. Recent advances in Shrimp aquaculture wastewater management. Heliyon 2021, 7, e08283. [Google Scholar] [CrossRef]
- Suzuki, A.; Nam, V.H. Better management practices and their outcomes in shrimp farming: Evidence from small-scale shrimp farmers in Southern Vietnam. Aquacult. Int. 2018, 26, 469–486. [Google Scholar] [CrossRef] [Green Version]
- Adams, A.; Thompson, K.D. Development of diagnostics for aquaculture: Challenges and opportunities. Aquac. Res. 2011, 42, 93–102. [Google Scholar] [CrossRef]
- Munang’andu, H.M.; Mugimba, K.K.; Byarugaba, D.K.; Mutoloki, S.; Evensen, Ø. Current advances on virus discovery and diagnostic role of viral metagenomics in aquatic organisms. Front. Microbiol. 2017, 8, 406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- OIE. Principles of validation of diagnostic assays for infectious diseases. In Manual of Diagnostic Tests for Aquatic Animals; Office International des E’pizooties: Paris, France, 2009. [Google Scholar]
- Sathish Kumar, T.; Navaneeth Krishnan, A.; Joseph Sahaya Rajan, J.; Makesh, M.; Jithendran, K.P.; Alavandi, S.V.; Vijayan, K.K. Visual loop-mediated isothermal amplification (LAMP) for the rapid diagnosis of Enterocytozoon hepatopenaei (EHP) infection. Parasitol. Res. 2018, 117, 1485–1493. [Google Scholar]
- Tsai, S.-K.; Chen, C.-C.; Lin, H.-J.; Lin, J.H.-J.; Chen, T.-T.; Wang, L.-C. Combination of multiplex reverse transcription recombinase polymerase amplification assay and capillary electrophoresis provides high sensitive and high-throughput simultaneous detection of avian influenza virus subtypes. J. Vet. Sci. 2020, 21, e24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sathish Kumar, T.; Radhika, K.; Joseph Sahaya Rajan, J.; Makesh, M.; Alavandi, S.V.; Vijayan, K.K. Closed-tube field-deployable loop-mediated isothermal amplification (LAMP) assay based on spore wall protein (SWP) for the visual detection of Enterocytozoon hepatopenaei (EHP). J. Invertebr. Pathol. 2021, 183, 107624. [Google Scholar] [CrossRef]
- Arunrut, N.; Kampeera, J.; Sirithammajak, S.; Sanguanrut, P.; Proespraiwong, P.; Suebsing, R.; Kiatpathomchai, W. Sensitive visual detection of AHPND bacteria using loop-mediated isothermal amplification combined with DNA-functionalized gold nanoparticles as probes. PLoS ONE 2016, 11, e0151769. [Google Scholar] [CrossRef] [PubMed]
- Mai, H.N.; Aranguren Caro, L.F.; Cruz-Flores, R.; Dhar, A.K. Development of a recombinase polymerase amplification (RPA) assay for acute hepatopancreatic necrosis disease (AHPND) detection in Pacific white shrimp (Penaeus vannamei). Mol. Cell. Probes 2021, 57, 101710. [Google Scholar] [CrossRef]
- Huang, Q.; Chen, Y.; Liu, H.; St-Hilaire, S.; Gao, S.; MacKinnon, B.; Zhu, S.; Wen, Z.; Jia, P.; Zheng, X. Establishment of a real-time Recombinase Polymerase Amplification (RPA) for the detection of decapod iridescent virus 1 (DIV1). J. Virol. Methods 2022, 300, 114377. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Fan, S.; Wang, Y.; Yang, H.; Qiao, Y.; Jiang, G.; Lyu, M.; Dong, J.; Shen, H.; Gao, S. Rapid detection of Enterocytozoon hepatopenaei infection in shrimp with a real-time isothermal recombinase polymerase amplification assay. Front. Cell Infect. Microbiol. 2021, 11, 631960. [Google Scholar] [CrossRef]
- Kanitchinda, S.; Srisala, J.; Suebsing, R.; Prachumwat, A.; Chaijarasphong, T. CRISPR-Cas fluorescent cleavage assay coupled with recombinase polymerase amplification for sensitive and specific detection of Enterocytozoon hepatopenaei. Biotechnol. Rep. 2020, 27, e00485. [Google Scholar] [CrossRef]
- Righetti, P.G.; Gelfi, C. Capillary electrophoresis of DNA in the 20–500 bp range: Recent developments. J. Biochem. Biophys. Methods 1999, 41, 75–90. [Google Scholar] [CrossRef]
- Slater, G.W.; Desruisseaux, C.; Hubert, S.J.; Mercier, J.F.; Labrie, J.; Boileau, J.; Tessier, F.; Pépin, M.P. Theory of DNA electrophoresis: A look at some current challenges. Electrophoresis 2000, 21, 3873–3887. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, X.; Ren, J. Recent advances in chemiluminescence detection coupled with capillary electrophoresis and microchip capillary electrophoresis. Electrophoresis 2016, 37, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Kerékgyártó, M.; Kerekes, T.; Tsai, E.; Amirkhanian, V.D.; Guttman, A. Light-emitting diode induced fluorescence (LED-IF) detection design for a pen-shaped cartridge based single capillary electrophoresis system. Electrophoresis 2012, 33, 2752–2758. [Google Scholar] [CrossRef] [PubMed]
- Cavanaugh, S.E.; Bathrick, A.S. Direct PCR amplification of forensic touch and other challenging DNA samples: A review. Forensic Sci. Int. Genet. 2018, 32, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors—Occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Ng, T.H.; Wang, H.C. Acute hepatopancreatic necrosis disease in penaeid shrimp. Rev. Aquac. 2020, 12, 1867–1880. [Google Scholar] [CrossRef] [Green Version]
- Jaroenlak, P.; Sanguanrut, P.; Williams, B.A.P.; Stentiford, G.D.; Flegel, T.W.; Sritunyalucksana, K.; Itsathitphaisarn, O. A nested PCR assay to avoid false positive detection of the microsporidian Enterocytozoon hepatopenaei (EHP) in environmental samples in shrimp farms. PLoS ONE 2016, 11, e0166320. [Google Scholar] [CrossRef] [Green Version]
- Mendoza-Cano, F.; Sánchez-Paz, A. Development and validation of a quantitative real-time polymerase chain assay for universal detection of the white spot syndrome virus in marine crustaceans. Virol. J. 2013, 10, 186. [Google Scholar] [CrossRef] [Green Version]
- Lai, H.-C.; Ng, T.-H.; Ando, M.; Lee, C.-T.; Chen, I.-T.; Chuang, J.-C.; Mavichak, R.; Chang, S.H.; Yeh, M.-D.; Chiang, Y.A.; et al. Pathogenesis of acute hepatopancreatic necrosis disease (AHPND) in shrimp. Fish Shellfish. Immunol. 2015, 47, 1006–1014. [Google Scholar] [CrossRef]
- Aranguren, L.F.; Han, J.E.; Tang, K.F.J. Enterocytozoon hepatopenaei (EHP) is a risk factor for acute hepatopancreatic necrosis disease (AHPND) and septic hepatopancreatic necrosis (SHPN) in the Pacific white shrimp Penaeus vannamei. Aquaculture 2017, 471, 37–42. [Google Scholar] [CrossRef] [Green Version]
- Chakrabarti, R.; Schutt, C.E. The enhancement of PCR amplification by low molecular weight amides. Nucleic Acids Res. 2001, 29, 2377–2381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winship, P.R. An improved method for directly sequencing PCR amplified material using dimethyl sulphoxide. Nucleic Acids Res. 1989, 17, 1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarkar, G.; Kapelner, S.; Sommer, S.S. Formamide can dramatically improve the specificity of PCR. Nucleic Acids Res. 1990, 18, 7465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, S.-K.; Shih, C.-H.; Chang, H.-W.; Teng, K.-H.; Hsu, W.-E.; Lin, H.-J.; Lin, H.-Y.; Huang, C.-H.; Chen, H.-W.; Wang, L.-C. Replication of a dog-origin H6N1 influenza virus in cell culture and mice. Viruses 2020, 12, 704. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-S.; Lo, C.-F.; Peng, S.-E.; Liu, K.-F.; Wang, C.-H.; Kou, G.-H. White spot syndrome virus (WSSV) PCR-positive Artemia cysts yield PCR-negative nauplii that fail to transmit WSSV when fed to shrimp postlarvae. Dis. Aquat. Org. 2002, 49, 1–10. [Google Scholar] [CrossRef]
- Tsai, Y.-L.; Wang, H.-C.; Lo, C.-F.; Tang-Nelson, K.; Lightner, D.; Ou, B.-R.; Hour, A.-L.; Tsai, C.-F.; Yen, C.-C.; Chang, G.H.-F.; et al. Validation of a commercial insulated isothermal PCR-based POCKIT test for rapid and easy detection of white spot syndrome virus infection in Litopenaeus vannamei. PLoS ONE 2014, 9, e90545. [Google Scholar] [CrossRef] [Green Version]
- Haq, M.B.; Priya, K.K.; Rajaram, R.; Vignesh, R.; Srinivasan, M. Real time PCR quantification of WSSV infection in specific pathogen free (SPF) Litopenaeus vannamei (Boone, 1931) exposed to antiviral nucleotide. Asian Pac. J. Trop. Biomed. 2012, 2, S1120–S1129. [Google Scholar] [CrossRef]
- Talukder, A.S.; Punom, N.J.; Eshik, M.M.E.; Begum, M.K.; Islam, H.M.R.; Hossain, Z.; Rahman, M.S. Molecular identification of white spot syndrome virus (WSSV) and associated risk factors for white spot disease (WSD) prevalence in shrimp (Penaeus monodon) aquaculture in Bangladesh. J. Invertebr. Pathol. 2021, 179, 107535. [Google Scholar] [CrossRef]
- Chaijarasphong, T.; Munkongwongsiri, N.; Stentiford, G.D.; Aldama-Cano, D.J.; Thansa, K.; Flegel, T.W.; Sritunyalucksana, K.; Itsathitphaisarn, O. The shrimp microsporidian Enterocytozoon hepatopenaei (EHP): Biology, pathology, diagnostics and control. J. Invertebr. Pathol. 2021, 186, 107458. [Google Scholar] [CrossRef]






| Disease | Target | Accession No. | Primer Name | DNA Sequence (5′-3′) | Anneal Temp. | Product Size | Reference |
|---|---|---|---|---|---|---|---|
| AHPND | Pir toxin gene | JALL01000066 | AP4-F2 | TTGAGAATACGGGACGTGGG | 55 °C | 230 bp | [29] |
| AP4-R2 | GTTAGTCATGTGAGCACCTTC | ||||||
| HPM | spore wall protein gene | KX258197 | SWP_2F | TTGGCGGCACAATTCTCAAACA | 64 °C | 148 bp | [30] |
| SWP_2R | GCTGTTTGTCTCCAACTGTATTTGA | ||||||
| WSD | VP28 envelope protein gene | AY249442 | VP28-140Fw | AGGTGTGGAACAACACATCAAG | 58 °C | 140 bp | [31] |
| VP28-140Rv | TGCCAACTTCATCCTCATCA |
| IQ2000 (Gold Standard) | ||||||
|---|---|---|---|---|---|---|
| Positive | Negative | Sensitivity (%) | Specificity (%) | Agreement (%) | ||
| DirectGO | Positive | 6 (TP) | 0 (FP) | 100 | 100 | 100 |
| Negative | 0 (FN) | 14 (TN) | ||||
| Procedure | IQ2000 | DirectGO |
|---|---|---|
| Pre-PCR | ||
| Sample treatment | ~50 min | ~20 min |
| PCR Premix | ~5 min | ~5 min |
| PCR | ||
| 1st PCR | 21 min | 50 min 20 s |
| PCR mature prepare | ~5 min | - |
| 2nd PCR | 30 min 10 s | - |
| Post-PCR | ||
| Make agarose gel | 30 min | - |
| Electrophoresis | 35 min | ~3 min |
| Gel stain and destain | 20 min | - |
| Data assay | 5 min | 1 min |
| Total time | >201 min | >86 min |
| Item | Direct PCR-Based CE | IQ2000 |
|---|---|---|
| Tissue weight | ~3 mg | ~20 mg |
| Total sample volume after extraction | 90 uL | 200 uL |
| Sample added per PCR reaction | 2.5 uL | 2 uL |
| Equivalent tissue weight per PCR reaction | ~83 ug | ~200 ug |
| Limitation of detection per PCR reaction (LOD) | 25–100 copies | 20 copies |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, H.-Y.; Yen, S.-C.; Tsai, S.-K.; Shen, F.; Lin, J.H.-Y.; Lin, H.-J. Combining Direct PCR Technology and Capillary Electrophoresis for an Easy-to-Operate and Highly Sensitive Infectious Disease Detection System for Shrimp. Life 2022, 12, 276. https://doi.org/10.3390/life12020276
Lin H-Y, Yen S-C, Tsai S-K, Shen F, Lin JH-Y, Lin H-J. Combining Direct PCR Technology and Capillary Electrophoresis for an Easy-to-Operate and Highly Sensitive Infectious Disease Detection System for Shrimp. Life. 2022; 12(2):276. https://doi.org/10.3390/life12020276
Chicago/Turabian StyleLin, Hung-Yun, Shao-Chieh Yen, Shou-Kuan Tsai, Fan Shen, John Han-You Lin, and Han-Jia Lin. 2022. "Combining Direct PCR Technology and Capillary Electrophoresis for an Easy-to-Operate and Highly Sensitive Infectious Disease Detection System for Shrimp" Life 12, no. 2: 276. https://doi.org/10.3390/life12020276