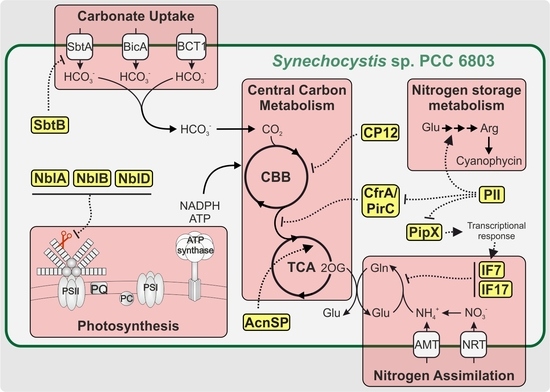
Small but Smart: On the Diverse Role of Small Proteins in the Regulation of Cyanobacterial Metabolism
Abstract
:1. Introduction
2. Light Regulation of the Calvin-Benson Cycle by the Small Protein CP12
3. Control of Glutamine Synthetase by Proteinaceous Inactivating Factors Unique to Cyanobacteria
4. Control of the Key Enzyme of Arginine Synthesis by Direct Interaction with the PII Protein
5. PII as an Antagonist for the Interaction of Small Proteins with Key Factors that Control Metabolic Fluxes
6. Further Examples of Small Protein Regulators Affecting the Activity of Enzymes or Transporters
7. The Potential of Small Proteins for Metabolic Engineering and Biotechnological Applications
Author Contributions
Funding
Conflicts of Interest
References
- Labeit, S.; Kolmerer, B. Titins: Giant proteins in charge of muscle ultrastructure and elasticity. Science 1995, 270, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Opitz, C.A.; Kulke, M.; Leake, M.C.; Neagoe, C.; Hinssen, H.; Hajjar, R.J.; Linke, W.A. Damped elastic recoil of the titin spring in myofibrils of human myocardium. Proc. Natl. Acad. Sci. USA 2003, 100, 12688–12693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galindo, M.I.; Pueyo, J.I.; Fouix, S.; Bishop, S.A.; Couso, J.P. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 2007, 5, 1052–1062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J. Protein-length distributions for the three domains of life. Trends Genet. 2000, 16, 107–109. [Google Scholar] [CrossRef]
- Miravet-Verde, S.; Ferrar, T.; Espadas-García, G.; Mazzolini, R.; Gharrab, A.; Sabido, E.; Serrano, L.; Lluch-Senar, M. Unraveling the hidden universe of small proteins in bacterial genomes. Mol. Syst. Biol. 2019, 15, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Storz, G.; Wolf, Y.I.; Ramamurthi, K.S. Small proteins can no Longer be ignored. Annu. Rev. Biochem. 2014, 83, 753–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmgren, A. Thioredoxin and glutaredoxin systems. Methods Enzymol. 1989, 264, 286–296. [Google Scholar] [CrossRef]
- Arnér, E.S.J.; Holmgren, A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2000, 267, 6102–6109. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Holmgren, A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014, 66, 75–87. [Google Scholar] [CrossRef]
- Su, M.; Ling, Y.; Yu, J.; Wu, J.; Xiao, J. Small proteins: Untapped area of potential biological importance. Front. Genet. 2013, 4, 286. [Google Scholar] [CrossRef]
- Kliemt, J.; Soppa, J. Diverse functions of small RNAs (sRNAs) in halophilic Archaea: From non-coding regulatory sRNAs to microprotein-Encoding sRNAs. In RNA Metabolism and Gene Expression in Archaea, 1st ed.; Clouet-d’Orval, B., Ed.; Springer: Dordrecht, The Netherlands, 2017; Volume 5, pp. 225–242. [Google Scholar]
- Grasse, N.; Mamedov, F.; Becker, K.; Styring, S.; Rögner, M.; Nowaczyk, M.M. Role of novel dimeric photosystem II (PSII)-Psb27 protein complex in PSII repair. J. Biol. Chem. 2011, 286, 29548–29555. [Google Scholar] [CrossRef] [Green Version]
- Rexroth, S.; Rexroth, D.; Veit, S.; Plohnke, N.; Cormann, K.U.; Nowaczyk, M.M.; Rögner, M. Functional characterization of the small regulatory subunit PetP from the cytochrome b6f complex in Thermosynechococcus elongatus. Plant Cell 2014, 26, 3435–3448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashino, Y.; Lauber, W.M.; Carroll, J.A.; Wang, Q.; Whitmarsh, J.; Satoh, K.; Pakrasi, H.B. Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 2002, 41, 8004–8012. [Google Scholar] [CrossRef] [PubMed]
- Guskov, A.; Kern, J.; Gabdulkhakov, A.; Broser, M.; Zouni, A.; Saenger, W. Cyanobacterial photosystem II at 2.9-Å resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 2009, 16, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, D.; Kopf, M.; Klähn, S.; Steglich, C.; Hess, W.R. Small proteins in cyanobacteria provide a paradigm for the functional analysis of the bacterial micro-proteome. BMC Microbiol. 2016, 16, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuber, P. A peptide profile of the Bacillus subtilis genome. Peptides 2001, 22, 1555–1577. [Google Scholar] [CrossRef]
- Yang, X.; Tschaplinski, T.J.; Hurst, G.B.; Jawdy, S.; Abraham, P.E.; Lankford, P.K.; Adams, R.M.; Shah, M.B.; Hettich, R.L.; Lindquist, E.; et al. Discovery and annotation of small proteins using genomics, proteomics, and computational approaches. Genome Res. 2011, 21, 634–641. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Schreiber, K.; Appel, J.; Makowka, A.; Fähnrich, B.; Roettger, M.; Hajirezaei, M.R.; Sönnichsen, F.D.; Schönheit, P.; Martin, W.F.; et al. The Entner-Doudoroff pathway is an overlooked glycolytic route in cyanobacteria and plants. Proc. Natl. Acad. Sci. USA 2016, 113, 5441–5446. [Google Scholar] [CrossRef] [Green Version]
- Stitt, M. Metabolic Regulation of Photosynthesis. In Photosynthesis and the Environment, 1st ed.; Baker, N.A., Ed.; Springer: Dordrecht, The Netherlands, 2006; Volume 5, pp. 151–190. [Google Scholar]
- Portis, A.R. Rubisco activase—Rubisco’s catalytic chaperone. Photosynth. Res. 2003, 75, 11–27. [Google Scholar] [CrossRef]
- Groben, R.; Kaloudas, D.; Raines, C.A.; Offmann, B.; Maberly, S.C.; Gontero, B. Comparative sequence analysis of CP12, a small protein involved in the formation of a Calvin cycle complex in photosynthetic organisms. Photosynth. Res. 2010, 103, 183–194. [Google Scholar] [CrossRef]
- Pohlmeyer, K.; Paap, B.K.; Soll, J.; Wedel, N. CP12: A small nuclear-encoded chloroplast protein provides novel insights into higher-plant GAPDH evolution. Plant Mol. Biol. 1996, 32, 969–978. [Google Scholar] [CrossRef]
- Wedel, N.; Soll, J.; Paap, B.K. CP12 provides a new mode of light regulation of Calvin cycle activity in higher plants. Proc. Natl. Acad. Sci. USA 1997, 94, 10479–10484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumura, H.; Kai, A.; Maeda, T.; Tamoi, M.; Satoh, A.; Tamura, H.; Hirose, M.; Ogawa, T.; Kizu, N.; Wadano, A.; et al. Structure basis for the regulation of glyceraldehyde-3-phosphate dehydrogenase activity via the intrinsically disordered protein CP12. Structure 2011, 19, 1846–1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reichmann, D.; Jakob, U. The roles of conditional disorder in redox proteins. Curr. Opin. Struct. Biol. 2013, 23, 436–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McFarlane, C.R.; Shah, N.R.; Kabasakal, B.V.; Echeverria, B.; Cotton, C.A.R.; Bubeck, D.; Murray, J.W. Structural basis of light-induced redox regulation in the Calvin–Benson cycle in cyanobacteria. Proc. Natl. Acad. Sci. USA 2019, 116, 20984–20990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graciet, E.; Lebreton, S.; Camadro, J.M.; Gontero, B. Characterization of native and recombinant A4 glyceraldehyde 3-phosphate dehydrogenase: Kinetic evidence for conformation changes upon association with the small protein CP12. Eur. J. Biochem. 2003, 270, 129–136. [Google Scholar] [CrossRef] [Green Version]
- Tamoi, M.; Miyazaki, T.; Fukamizo, T.; Shigeoka, S. The Calvin cycle in cyanobacteria is regulated by CP12 via the NAD(H)/NADP(H) ratio under light/dark conditions. Plant J. 2005, 42, 504–513. [Google Scholar] [CrossRef]
- Thompson, L.R.; Zeng, Q.; Kelly, L.; Huang, K.H.; Singer, A.U.; Stubbe, J.A.; Chisholm, S.W. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc. Natl. Acad. Sci. USA 2011, 108. [Google Scholar] [CrossRef] [Green Version]
- Esteves-Ferreira, A.A.; Cavalcanti, J.H.F.; Vaz, M.G.M.V.; Alvarenga, L.V.; Nunes-Nesi, A.; Araújo, W.L. Cyanobacterial nitrogenases: Phylogenetic diversity, regulation and functional predictions. Genet. Mol. Biol. 2017, 40, 261–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zehr, J.P. Nitrogen fixation by marine cyanobacteria. Trends Microbiol. 2011, 19, 162–173. [Google Scholar] [CrossRef]
- Quintero, M.J.; Montesinos, M.L.; Herrero, A.; Flores, E. Identification of genes encoding amino acid permeases by inactivation of selected ORFs from the Synechocystis genomic sequence. Genome Res. 2001, 11, 2034–2040. [Google Scholar] [CrossRef] [Green Version]
- Valladares, A.; Montesinos, M.L.; Herrero, A.; Flores, E. An ABC-type, high-affinity urea permease identified in cyanobacteria. Mol. Microbiol. 2002, 43, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Flores, E.; Frías, J.E.; Rubio, L.M.; Herrero, A. Photosynthetic nitrate assimilation in cyanobacteria. Photosynth. Res. 2005, 83, 117–133. [Google Scholar] [CrossRef]
- Muro-Pastor, M.I.; Reyes, J.C.; Florencio, F.J. Ammonium assimilation in cyanobacteria. Photosynth. Res. 2005, 83, 135–150. [Google Scholar] [CrossRef] [PubMed]
- Kamennaya, N.A.; Chernihovsky, M.; Post, A.F. The cyanate utilization capacity of marine unicellular Cyanobacteria. Limnol. Oceanogr. 2008, 53, 2485–2494. [Google Scholar] [CrossRef] [Green Version]
- Forchhammer, K.; Tandeau De Marsac, N. The P(II) protein in the cyanobacterium Synechococcus sp. strain PCC 7942 is modified by serine phosphorylation and signals the cellular N-status. J. Bacteriol. 1994, 176, 84–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merrick, M.J.; Edwards, R.A. Nitrogen control in bacteria. Microbiol. Rev. 1995, 59, 604–622. [Google Scholar] [CrossRef] [Green Version]
- García-Domínguez, M.; Reyes, J.C.; Florencio, F.J. Glutamine synthetase inactivation by protein-protein interaction. Proc. Natl. Acad. Sci. USA 1999, 96, 7161–7166. [Google Scholar] [CrossRef] [Green Version]
- García-Domínguez, M.; Reyes, J.C.; Florencio, F.J. NtcA represses transcription of gifA and gifB, genes that encode inhibitors of glutamine synthetase type I from Synechocystis sp. PCC 6803. Mol. Microbiol. 2000, 35, 1192–1201. [Google Scholar] [CrossRef] [Green Version]
- Herrero, A.; Muro-Pastor, A.M.; Flores, E. Nitrogen control in cyanobacteria. J. Bacteriol. 2001, 183, 411–425. [Google Scholar] [CrossRef] [Green Version]
- Klähn, S.; Schaal, C.; Georg, J.; Baumgartner, D.; Knippen, G.; Hagemann, M.; Muro-Pastor, A.M.; Hess, W.R. The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc. Natl. Acad. Sci. USA 2015, 112, E6243–E6252. [Google Scholar] [CrossRef] [Green Version]
- Klähn, S.; Bolay, P.; Wright, P.R.; Atilho, R.M.; Brewer, K.I.; Hagemann, M.; Breaker, R.R.; Hess, W.R. A glutamine riboswitch is a key element for the regulation of glutamine synthetase in cyanobacteria. Nucleic Acids Res. 2018, 46, 10082–10094. [Google Scholar] [CrossRef] [Green Version]
- Bolay, P.; Muro-Pastor, M.I.; Florencio, F.J.; Klähn, S. The distinctive regulation of cyanobacterial glutamine synthetase. Life 2018, 8, 52. [Google Scholar] [CrossRef] [Green Version]
- Gutt, M.; Jordan, B.; Weidenbach, K.; Gudzuhn, M.; Kiessling, C.; Cassidy, L.; Helbig, A.; Tholey, A.; Pyper, D.; Schwalbe, H.; et al. Small protein 26 interacts and enhances glutamine synthetase activity in Methanosarcina mazei. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ehlers, C.; Weidenbach, K.; Veit, K.; Forchhammer, K.; Schmitz, R.A. Unique mechanistic features of post-translational regulation of glutamine synthetase activity in Methanosarcina mazei strain Gö1 in response to nitrogen availability. Mol. Microbiol. 2005, 55, 1841–1854. [Google Scholar] [CrossRef]
- Huergo, L.F.; Chandra, G.; Merrick, M. PII signal transduction proteins: Nitrogen regulation and beyond. FEMS Microbiol. Rev. 2013, 37, 251–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.M.; Flores, E.; Herrero, A.; Houmard, J.; Tandeau De Marsac, N. A role for the signal transduction protein P(II) in the control of nitrate/nitrite uptake in a cyanobacterium. FEBS Lett. 1998, 427, 291–295. [Google Scholar] [CrossRef] [Green Version]
- Heinrich, A.; Maheswaran, M.; Ruppert, U.; Forchhammer, K. The Synechococcus elongatus PII signal transduction protein controls arginine synthesis by complex formation with N-acetyl-L-glutamate kinase. Mol. Microbiol. 2004, 52, 1303–1314. [Google Scholar] [CrossRef]
- Simon, R.D. Cyanophycin Granules from the Blue-Green Alga Anabaena cylindrica: A Reserve Material Consisting of Copolymers of Aspartic Acid and Arginine. Proc. Natl. Acad. Sci. USA 1971, 68, 265–267. [Google Scholar] [CrossRef] [Green Version]
- Pauwels, K.; Abadjieva, A.; Hilven, P.; Stankiewicz, A.; Crabeel, M. The N-acetylglutamate synthase/N-acetylglutamate kinase metabolon of Saccharomyces cerevisiae allows coordinated feedback regulation of the first two steps in arginine biosynthesis. Eur. J. Biochem. 2003, 270, 1014–1024. [Google Scholar] [CrossRef] [Green Version]
- Caldovic, L.; Tuchman, M. N -Acetylglutamate and its changing role through evolution. Biochem J. 2003, 290, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Cunin, R.; Glansdorff, N.; Pierard, A.; Stalon, V. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 1986, 50, 314–352. [Google Scholar] [CrossRef] [PubMed]
- Maheswaran, M.; Urbanke, C.; Forchhammer, K. Complex formation and catalytic activation by the PII signaling protein of N-acetyl-L-glutamate kinase from Synechococcus elongatus strain PCC 7942. J. Biol. Chem. 2004, 279, 55202–55210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forchhammer, K. PII signal transducers: Novel functional and structural insights. Trends Microbiol. 2008, 16, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Maheswaran, M.; Forchhammer, K. Carbon-source-dependent nitrogen regulation in Escherichia coli is mediated through glutamine-dependent GlnB signalling. Microbiology 2003, 149, 2163–2172. [Google Scholar] [CrossRef] [PubMed]
- Forchhammer, K.; Tandeau De Marsac, N. Functional analysis of the phosphoprotein P(II) (glnB gene product) in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 1995, 177, 2033–2040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forchhammer, K. Global carbon/nitrogen control by PII signal transduction in cyanobacteria: From signals to targets. FEMS Microbiol. Rev. 2004, 28, 319–333. [Google Scholar] [CrossRef] [Green Version]
- Forchhammer, K. The PII protein in Synechococcus PCC 7942 senses and signals 2-oxoglutarate under ATP-replete conditions. In The Phototrophic Prokaryotes, 1st ed.; Peschek, G.A., Löffelhardt, W., Schmetterer, G., Eds.; Springer: Berlin/Heidelberg, Germany, 1999; pp. 549–553. [Google Scholar]
- Forchhammer, K.; De Marsac, N.T. Phosphorylation of the P(II) protein (glnB gene product) in the cyanobacterium Synechococcus sp. strain PCC 7942: Analysis of in vitro kinase activity. J. Bacteriol. 1995, 177, 5812–5817. [Google Scholar] [CrossRef] [Green Version]
- Kloft, N.; Rasch, G.; Forchhammer, K. Protein phosphatase PphA from Synechocystis sp. PCC 6803: The physiological framework of PII-P dephosphorylation. Microbiology 2005, 151, 1275–1283. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.M.; Flores, E.; Forchhammer, K.; Herrero, A.; Tandeau De Marsac, N. Phosphorylation of the signal transducer P(II) protein and an additional effector are required for the P(II)-mediated regulation of nitrate and nitrite uptake in the cyanobacterium Synechococcus sp. PCC 7942. Eur. J. Biochem. 2000, 267, 591–600. [Google Scholar] [CrossRef] [Green Version]
- Heinrich, A.; Woyda, K.; Brauburger, K.; Meiss, G.; Detsch, C.; Stülke, J.; Forchhammer, K. Interaction of the membrane-bound GlnK-AmtB complex with the master regulator of nitrogen metabolism TnrA in Bacillus subtilis. J. Biol. Chem. 2006, 281, 34909–34917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radchenko, M.V.; Thornton, J.; Merrick, M. Control of AmtB-GlnK complex formation by intracellular levels of ATP, ADP, and 2-oxoglutarate. J. Biol. Chem. 2010, 285, 31037–31045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watzer, B.; Spät, P.; Neumann, N.; Koch, M.; Sobotka, R.; MacEk, B.; Hennrich, O.; Forchhammer, K. The signal transduction protein PII controls ammonium, nitrate and urea uptake in cyanobacteria. Front. Microbiol. 2019, 10, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hisbergues, M.; Jeanjean, R.; Joset, F.; Tandeau De Marsac, N.; Bédu, S. Protein PII regulates both inorganic carbon and nitrate uptake and is modified by a redox signal in Synechocystis PCC 6803. FEBS Lett. 1999, 463, 216–220. [Google Scholar] [CrossRef] [Green Version]
- Hauf, W.; Schmid, K.; Gerhardt, E.C.M.; Huergo, L.F.; Forchhammer, K. Interaction of the nitrogen regulatory protein GlnB (PII) with biotin carboxyl carrier protein (BCCP) controls acetyl-CoA levels in the cyanobacterium Synechocystis sp. PCC 6803. Front. Microbiol. 2016, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Osanai, T.; Sato, S.; Tabata, S.; Tanaka, K. Identification of PamA as a PII-binding membrane protein important in nitrogen-related and sugar-catabolic gene expression in Synechocystis sp. PCC 6803. J. Biol. Chem. 2005, 280, 34684–34690. [Google Scholar] [CrossRef] [Green Version]
- Muro-Pastor, M.I.; Cutillas-Farray, Á.; Pérez-Rodríguez, L.; Pérez-Saavedra, J.; Vega-de Armas, A.; Paredes, A.; Robles-Rengel, R.; Florencio, F.J. CfrA, a novel carbon flow regulator, adapts carbon metabolism to nitrogen deficiency in cyanobacteria. Plant Physiol. 2020. [Google Scholar] [CrossRef]
- Orthwein, T.; Scholl, J.; Spät, P.; Lucius, S.; Koch, M.; Macek, B.; Hagemann, M.; Forchhammer, K. The Novel PII-Interacting Regulator PirC (Sll0944) Identifies 3-Phosphoglycerate Mutase (PGAM) as Central Control Point of Carbon Storage Metabolism in Cyanobacteria. bioRxiv 2020. [Google Scholar] [CrossRef]
- Leigh, J.A.; Dodsworth, J.A. Nitrogen regulation in bacteria and archaea. Annu. Rev. Microbiol. 2007, 61, 349–377. [Google Scholar] [CrossRef]
- Espinosa, J.; Forchhammer, K.; Burillo, S.; Contreras, A. Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol. Microbiol. 2006, 61, 457–469. [Google Scholar] [CrossRef]
- Giner-Lamia, J.; Robles-Rengel, R.; Hernández-Prieto, M.A.; Isabel Muro-Pastor, M.; Florencio, F.J.; Futschik, M.E. Identification of the direct regulon of NtcA during early acclimation to nitrogen starvation in the cyanobacterium Synechocystis sp. PCC 6803. Nucleic Acids Res. 2017, 45, 11800–11820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llácer, J.L.; Espinosa, J.; Castells, M.A.; Contreras, A.; Forchhammer, K.; Rubio, V. Structural basis for the regulation of NtcA-dependent transcription by proteins PipX and PII. Proc. Natl. Acad. Sci. USA 2010, 107, 15397–15402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, M.-X.; Jiang, Y.-L.; He, Y.-X.; Chen, Y.-F.; Teng, Y.-B.; Chen, Y.; Zhang, C.-C.; Zhou, C.-Z. Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate. Proc. Natl. Acad. Sci. USA 2010, 107, 12487–12492. [Google Scholar] [CrossRef] [Green Version]
- Forcada-Nadal, A.; Llácer, J.L.; Contreras, A.; Marco-Marín, C.; Rubio, V. The PII-NAGK-PipX-NtcA regulatory axis of cyanobacteria: A tale of changing partners, allosteric effectors and non-covalent interactions. Front. Mol. Biosci. 2018, 5, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Labella, J.I.; Obrebska, A.; Espinosa, J.; Salinas, P.; Forcada-Nadal, A.; Tremiño, L.; Rubio, V.; Contreras, A. Expanding the cyanobacterial nitrogen regulatory network: The GntR-like regulator PlmA interacts with the PII-PipX complex. Front. Microbiol. 2016, 7, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Labella, J.I.; Cantos, R.; Salinas, P.; Espinosa, J.; Contreras, A. Distinctive features of PipX, a unique signaling protein of cyanobacteria. Life 2020, 10, 79. [Google Scholar] [CrossRef] [PubMed]
- Bolay, P.; Muro-pastor, M.I.; Rozbeh, R.; Timm, S.; Hagemann, M.; Florencio, F.J.; Forchhammer, K.; Klähn, S. The novel PII-interacting protein PirA regulates flux into the cyanobacterial ornithine-ammonia cycle. bioRxiv 2020. [Google Scholar] [CrossRef]
- Forchhammer, K.; Lüddecke, J. Sensory properties of the PII signalling protein family. FEBS J. 2016, 283, 425–437. [Google Scholar] [CrossRef]
- Van Heeswijk, W.C.; Hoving, S.; Molenaar, D.; Stegeman, B.; Kahn, D.; Westerhoff, H.V. An alternative P(II) protein in the regulation of glutamine synthetase in Escherichia coli. Mol. Microbiol. 1996, 21, 133–146. [Google Scholar] [CrossRef] [Green Version]
- Atkinson, M.R.; Ninfa, A.J. Role of the GlnK signal transduction protein in the regulation of nitrogen assimilation in Escherichia coli. Mol. Microbiol. 1998, 29, 431–447. [Google Scholar] [CrossRef] [Green Version]
- Du, J.; Förster, B.; Rourke, L.; Howitt, S.M.; Price, G.D. Characterisation of cyanobacterial bicarbonate transporters in E. coli shows that SbtA homologs are functional in this heterologous expression system. PLoS ONE 2014, 9, e115905. [Google Scholar] [CrossRef] [PubMed]
- Selim, K.A.; Haase, F.; Hartmann, M.D.; Hagemann, M.; Forchhammer, K. PII-like signaling protein SbtB links cAMP sensing with cyanobacterial inorganic carbon response. Proc. Natl. Acad. Sci. USA 2018, 115, E4861–E4869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaczmarski, J.A.; Hong, N.S.; Mukherjee, B.; Wey, L.T.; Rourke, L.; Förster, B.; Peat, T.S.; Price, G.D.; Jackson, C.J. Structural basis for the allosteric regulation of the SbtA bicarbonate transporter by the PII-like protein, SbtB, from Cyanobium sp. PCC7001. Biochemistry 2019, 58, 5030–5039. [Google Scholar] [CrossRef] [PubMed]
- de Alvarenga, L.V.; Hess, W.R.; Hagemann, M. AcnSP—A novel small protein regulator of aconitase activity in the cyanobacterium Synechocystis sp. PCC 6803. Front. Microbiol. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Collier, J.L.; Grossman, A.R. A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 1994, 13, 1039–1047. [Google Scholar] [CrossRef] [PubMed]
- Bienert, R.; Baier, K.; Volkmer, R.; Lockau, W.; Heinemann, U. Crystal structure of NblA from Anabaena sp. PCC 7120, a small protein playing a key role in phycobilisome degradation. J. Biol. Chem. 2006, 281, 5216–5223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karradt, A.; Sobanski, J.; Mattow, J.; Lockau, W.; Baier, K. NblA, a key protein of phycobilisome degradation, interacts with ClpC, a HSP100 chaperone partner of a cyanobacterial Clp protease. J. Biol. Chem. 2008, 283, 32394–32403. [Google Scholar] [CrossRef] [Green Version]
- Dolganov, N.; Grossman, A.R. A polypeptide with similarity to phycocyanin α-subunit phycocyanobilin lyase involved in degradation of phycobilisomes. J. Bacteriol. 1999, 181, 610–617. [Google Scholar] [CrossRef] [Green Version]
- Hu, P.P.; Hou, J.Y.; Xu, Y.L.; Niu, N.N.; Zhao, C.; Lu, L.; Zhou, M.; Scheer, H.; Zhao, K.H. The role of lyases, NblA and NblB proteins and bilin chromophore transfer in restructuring the cyanobacterial light-harvesting complex. Plant J. 2020, 102, 529–540. [Google Scholar] [CrossRef]
- Krauspe, V.; Fahrner, M.; Spät, P.; Steglich, C.; Frankenberg-Dinkel, N.; Macek, B.; Schilling, O.; Hess, W.R. Discovery of a novel small protein factor involved in the coordinated degradation of phycobilisomes in cyanobacteria. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kopf, M.; Klähn, S.; Scholz, I.; Matthiessen, J.K.F.; Hess, W.R.; Voß, B. Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res. 2014, 21, 527–539. [Google Scholar] [CrossRef]
- Hitchcock, A.; Hunter, C.N.; Canniffe, D.P. Progress and challenges in engineering cyanobacteria as chassis for light-driven biotechnology. Microb. Biotechnol. 2020, 13, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Savakis, P.; Hellingwerf, K.J. Engineering cyanobacteria for direct biofuel production from CO2. Curr. Opin. Biotechnol. 2015, 33, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Hagemann, M.; Hess, W.R. Systems and synthetic biology for the biotechnological application of cyanobacteria. Curr. Opin. Biotechnol. 2018, 49, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Ducat, D.C.; Way, J.C.; Silver, P.A. Engineering cyanobacteria to generate high-value products. Trends Biotechnol. 2011, 29, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Abed, R.M.M.; Dobretsov, S.; Sudesh, K. Applications of cyanobacteria in biotechnology. J. Appl. Microbiol. 2009, 106, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rosgaard, L.; de Porcellinis, A.J.; Jacobsen, J.H.; Frigaard, N.U.; Sakuragi, Y. Bioengineering of carbon fixation, biofuels, and biochemicals in cyanobacteria and plants. J. Biotechnol. 2012, 162, 134–147. [Google Scholar] [CrossRef] [PubMed]
- David, C.; Schmid, A.; Adrian, L.; Wilde, A.; Bühler, K. Production of 1,2-propanediol in photoautotrophic Synechocystis is linked to glycogen turn-over. Biotechnol. Bioeng. 2018, 115, 300–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoschek, A.; Toepel, J.; Hochkeppel, A.; Karande, R.; Bühler, B.; Schmid, A. Light-dependent and aeration-independent gram-scale hydroxylation of cyclohexane to cyclohexanol by CYP450 harboring Synechocystis sp. PCC 6803. Biotechnol. J. 2019, 14, 1–10. [Google Scholar] [CrossRef]
- Van Dam, J.E.G.; De Klerk-Engels, B.; Struik, P.C.; Rabbinge, R. Securing renewable resource supplies for changing market demands in a bio-based economy. Ind. Crops Prod. 2005, 21, 129–144. [Google Scholar] [CrossRef]
- Wang, B.; Wang, J.; Meldrum, D.R. Application of synthetic biology in cyanobacteria and algae. Front. Microbiol. 2012, 3, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markley, A.L.; Begemann, M.B.; Clarke, R.E.; Gordon, G.C.; Pfleger, B.F. Synthetic biology toolbox for controlling gene expression in the cyanobacterium Synechococcus sp. strain PCC 7002. ACS Synth. Biol. 2015, 4, 595–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Englund, E.; Liang, F.; Lindberg, P. Evaluation of promoters and ribosome binding sites for biotechnological applications in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Zhou, J.; Zhu, T.; Cai, Z.; Li, Y. From cyanochemicals to cyanofactories: A review and perspective. Microb. Cell Fact. 2016, 15, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gründel, M.; Scheunemann, R.; Lockau, W.; Zilliges, Y. Impaired glycogen synthesis causes metabolic overflow reactions and affects stress responses in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 2012, 158, 3032–3043. [Google Scholar] [CrossRef]
- Srivastava, A.; Brilisauer, K.; Rai, A.K.; Ballal, A.; Forchhammer, K.; Tripathi, A.K. Down-regulation of the alternative sigma factor SIGJ confers a photoprotective phenotype to Anabaena PCC 7120. Plant Cell Physiol. 2017, 58, 287–297. [Google Scholar] [CrossRef]
- Tailor, V.; Ballal, A. Novel molecular insights into the function and the antioxidative stress response of a Peroxiredoxin Q protein from cyanobacteria. Free Radic. Biol. Med. 2017, 106, 278–287. [Google Scholar] [CrossRef]
- Frizzi, A.; Huang, S. Tapping RNA silencing pathways for plant biotechnology. Plant Biotechnol. J. 2010, 8, 655–677. [Google Scholar] [CrossRef]
- Kanno, M.; Carroll, A.L.; Atsumi, S. Global metabolic rewiring for improved CO2 fixation and chemical production in cyanobacteria. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Koch, M.; Bruckmoser, J.; Scholl, J.; Hauf, W.; Rieger, B.; Forchhammer, K. Maximizing PHB content in Synechocystis sp. PCC 6803: Development of a new photosynthetic overproduction strain. bioRxiv 2020. [Google Scholar] [CrossRef]
- Watzer, B.; Engelbrecht, A.; Hauf, W.; Stahl, M.; Maldener, I.; Forchhammer, K. Metabolic pathway engineering using the central signal processor PII. Microb. Cell Fact. 2015, 14, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brandenburg, F.; Klähn, S. Small but Smart: On the Diverse Role of Small Proteins in the Regulation of Cyanobacterial Metabolism. Life 2020, 10, 322. https://doi.org/10.3390/life10120322
Brandenburg F, Klähn S. Small but Smart: On the Diverse Role of Small Proteins in the Regulation of Cyanobacterial Metabolism. Life. 2020; 10(12):322. https://doi.org/10.3390/life10120322
Chicago/Turabian StyleBrandenburg, Fabian, and Stephan Klähn. 2020. "Small but Smart: On the Diverse Role of Small Proteins in the Regulation of Cyanobacterial Metabolism" Life 10, no. 12: 322. https://doi.org/10.3390/life10120322