Riemann-Symmetric-Space-Based Models in Screening for Gene Transfer Polymers
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mary, B.; Maurya, S.; Kumar, M.; Bammidi, S.; Kumar, V.; Jayandharan, G.R. Molecular engineering of Adeno-associated virus capsid improves its therapeutic gene transfer in murine models of hemophilia and retinal degeneration. Mol. Pharm. 2019, 16, 4738–4750. [Google Scholar] [CrossRef] [PubMed]
- Aartsma-Rus, A.; van Putten, M. The use of genetically humanized animal models for personalized medicine approaches. Dis. Model. Mech. 2019, 13, 041673. [Google Scholar] [CrossRef] [PubMed]
- Contin, M.; Garcia, C.; Dobrecky, C.; Lucangioli, S.; D’Accorso, N. Advances in drug delivery, gene delivery and therapeutic agents based on dendritic materials. Future Med. Chem. 2019, 11, 1791–1810. [Google Scholar] [CrossRef] [PubMed]
- Richard, B.; Ho-Fung, C.; Jóhannes, R. Characteristics of known drug space. Natural products, their derivatives and synthetic drugs. Eur. J. Med. Chem. 2010, 45, 5646–5652. [Google Scholar]
- Anderson, D.G.; Peng, W.; Akinc, A.; Naushad, H.; Kohn, A.; Pandera, R.; Langer, R.; Sawicli, A.J. A polymer library approach to suicide gene therapy for cancer. Proc. Natl. Acad. Sci. USA 2004, 9, 16028–16033. [Google Scholar] [CrossRef] [PubMed]
- Beata, S.; Mircea, V.D.; Mihai, V.P.; Ireneusz, P.G. Docking linear ligands to glucose oxidase. Int. J. Mol. Sci. 2016, 17, 1796. [Google Scholar]
- Nguyen, L.H.; Nguyen, T.H.; Truong, T.N. Quantum Mechanical-Based Quantitative Structure-Property Relationships for Electronic Properties of Two Large Classes of Organic Semiconductor Materials: Polycyclic Aromatic Hydrocarbons and Thienoacenes. ACS Omega 2019, 4, 7516–7523. [Google Scholar] [CrossRef] [PubMed]
- Mathematica; Version 8.0; Wolfram Research, Inc.: Champaign, IL, USA, 2010.
- Schrödinger Release Prime; Schrödinger LLC: New York, NY, USA, 2009.
- Akhiezer, D.N.; Vinberg, E.B. Weakly symmetric spaces and spherical varieties. Transf. Groups 1999, 4, 3–24. [Google Scholar] [CrossRef]
- Wolf Joseph, A. Spaces of Constant Curvature, 5th ed.; McGraw–Hill: New York, NY, USA, 1999. [Google Scholar]
- Conway John, B. Functions of One Complex Variable, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 1978. [Google Scholar]
- Arfken, G. Mathematical Methods for Physicists, 3rd ed.; Academic Press: Orlando, FL, USA, 1985; pp. 397–399. [Google Scholar]
- Morse, P.M.; Feshbach, H. Methods of Theoretical Physics, Part I; McGraw-Hill: New York, NY, USA, 1953; pp. 391–392, 399–401. [Google Scholar]
- Trott, M. The Mathematica Guidebook for Programming; Springer: Berlin/Heidelberg, Germany, 2004; pp. 188–191. [Google Scholar]
- Weisstein, E.W. “Branch Point” from MathWorld—A WolframWeb Resource. Available online: http://mathworld.wolfram.com/BranchPoint.html (accessed on 5 December 2018).
- Majumdar, S.; Basak, S.C.; Lungu, C.N.; Diudea, M.V.; Grunwald, G.D. Mathematical structural descriptors and mutagenicity assessment: A study with congeneric and diverse datasets. SAR QSAR Environ. Res. 2018, 29, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Petitjean, M. A definition of symmetry. Symmetry Cult. Sci. 2007, 18, 99–119. [Google Scholar]
- Lungu, C.N. C-C chemokine receptor type 3 inhibitors: Bioactivity prediction using local vertex invariants based on thermal conductivity layer matrix. Stud. Univ. Babes-Bolyai Chem. 2018, 63, 177–188. [Google Scholar] [CrossRef]
- Lungu, C.N.; Diudea, M.V.; Putz, M.V. Ligand shaping in induced fit docking of MraY inhibitors. Polynomial discriminant and Laplacian operator as biological activity descriptors. Int. J. Mol. Sci. 2018, 18, 1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Item | Monomers with Proven Gene Transfer Capabilities |
|---|---|
| 1 |  |
| 2 |  |
| 3 |  |
| 4 |  |
| 5 |  |
| 6 |  |
| 7 |  |
| 8 |  |
| 9 |  |
| 10 |  |
| 11 |  |
| 11 |  |
| 12 |  |
| 13 |  |
| 14 |  |
| 15 |  |
| 16 |  |
| 17 |  |
| 18 |  |
| 19 |  |
| 20 |  |
| 21 |  |
| 22 |  |
| 23 |  |
| 24 |  |
| 25 |  |
| 26 |  |
| 27 |  |
| 28 |  |
| 29 |  |
| Monomer Equations | |
|---|---|
| # | Coordinate Based Equations |
| 1 | ∂H(−13.4 + 19H) |
| 2 | ∂H(−2.2 + 18.5H) |
| 3 | ∂H(−13.0 + 28H) |
| 4 | ∂H(−13.6 + 14H) |
| 5 | ∂H(−2.4 + 15H) |
| 6 | ∂H(−2.6 + 15H) |
| 7 | ∂H(−4.9 + 17H) |
| 8 | ∂H(−15.5 + 20H) |
| 9 | ∂H(−4.5 + 23H) |
| 10 | ∂H(−4.3 + 18H) |
| 11 | ∂H(−19.1 + 24H) |
| 12 | ∂H(−3.4 + 19H) |
| 13 | ∂H(−3.7 + 24H) |
| 14 | ∂H(−2.4 + 21H) |
| 15 | ∂H(−4.6 + 26H) |
| 16 | ∂H(−4.0 + 2.8H) |
| 17 | ∂H(6.8H) |
| 18 | ∂H(−1.8 + 19H) |
| 19 | ∂H(−1.9 + 795.2H) |
| 20 | ∂H(−13.4 + 28H) |
| 21 | ∂H(−28.7 + 28H) |
| 22 | ∂H(−16.7 + 59H) |
| 23 | ∂H(83.1H) |
| 24 | ∂H(−10.8 + 31H) |
| 25 | ∂H(25H) |
| 26 | ∂H(31H) |
| 27 | ∂H(−7.3 + 323.5H) |
| 28 | ∂H(398.6H) |
| 29 | ∂H(−5.8 + 49H) |
| # | Equations for Computing Riemann Surfaces |
|---|---|
| 1 | z (−13.4 + 19 z)^(1/(z (−13.4 + 19 z))) |
| 2 | z (−2.2 + 18.5z)^(1/(z (−2.2 + 18.5z))) |
| 3 | z (−13.0 + 28z)^(1/(z (−13.0 + 28z))) |
| 4 | z (−13.6 + 14 z)^(1/(z (−13.6 + 14z))) |
| 5 | z (−2.4 + 15 z)^(1/(z (−2.4 + 15 z))) |
| 6 | z (−2.6 + 15z)^(1/(z (−2.6 + 15z))) |
| 7 | z (−4.9 + 17z)^(1/(z (−4.9 + 17z))) |
| 8 | z (−15.5 + 20z)^(1/(z (−15.5 + 20z))) |
| 9 | z (−4.5 + 23z)^(1/(z (−4.5 + 23 z))) |
| 10 | z (−4.3 + 18z)^(1/(z (−4.3 + 18z))) |
| 11 | z (−19.1 + 24z)^(1/(z (−19.1 + 24z))) |
| 12 | z (−3.4 + 19 z)^(1/(z (−3.4 + 19z))) |
| 13 | z (−3.7 + 24 z)^(1/(z (−3.7 + 24z))) |
| 14 | z (−2.4 + 21z)^(1/(−2.4 + 21 z))) |
| 15 | z (−4.6 + 26z)^(1/(z (−4.6 + 26 z))) |
| 16 | z (−4.0 + 2.8 z)^(1/(z (−4.0 + 28 z))) |
| 17 | z (6.8 z)^(1/(z (6.8 z))) |
| 18 | z (−1.8 + 19z)^(1/(z (−1.8 + 19 z))) |
| 19 | z (−1.9 + 795.2 z)^(1/(z (−1.9 + 795.2 z))) |
| 20 | z (−13.4 + 28 z)^(1/(z (−13.4 + 28 z))) |
| 21 | z (−28.7 + 28 z)^(1/(z (−28.7 + 28 z))) |
| 22 | z (−16.7 + 59 z)^(1/(z (−16.7 + 59 z))) |
| 23 | z ( 83.1 z)^(1/(z (83.1 z))) |
| 24 | z (−10.8 + 31 z)^(1/(z (−10.8 + 31 z))) |
| 25 | z (25 z)^(1/(z ( 25 z))) |
| 26 | z (31 z)^(1/(z ( 31z))) |
| 27 | z (−7.3 + 323.5 z)^(1/(z (−7.3 + 323.5 z))) |
| 28 | z ( 398.6 z)^(1/(z (398.6 z))) |
| 29 | z (−5.8 + 49 z)^(1/(z (−5.8 + 49 z))) |
| Riemann Surfaces | ||
|---|---|---|
| 1 |  |  |
| 2 |  |  |
| 3 |  |  |
| 4 |  |  |
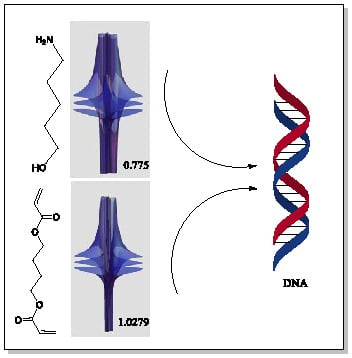
| 5 | 0.7754 | 1.027 |
| 6 | # 8 | # 21 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lungu, C.N.; Grudzinski, I.P. Riemann-Symmetric-Space-Based Models in Screening for Gene Transfer Polymers. Symmetry 2019, 11, 1466. https://doi.org/10.3390/sym11121466
Lungu CN, Grudzinski IP. Riemann-Symmetric-Space-Based Models in Screening for Gene Transfer Polymers. Symmetry. 2019; 11(12):1466. https://doi.org/10.3390/sym11121466
Chicago/Turabian StyleLungu, Claudiu N., and Ireneusz P. Grudzinski. 2019. "Riemann-Symmetric-Space-Based Models in Screening for Gene Transfer Polymers" Symmetry 11, no. 12: 1466. https://doi.org/10.3390/sym11121466