Rare Deletions or Large Duplications Contribute to Genetic Variation in Patients with Severe Tinnitus and Meniere Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Selection
2.2. DNA Sequencing and Dataset Generation
2.3. Variant Prioritization
2.4. Public Database Annotation
- RNA-Seq in human brain tissues: Amygdala, Anterior cingulate cortex (BA24), Caudate (basal ganglia), Cerebellar Hemisphere, Cerebellum, Cortex, Frontal Cortex (BA9), Hippocampus, Hypothalamus, Nucleus accumbens (basal ganglia), Putamen (basal ganglia), Spiral cord (cervical c-1), and Substantia nigra. The gene Transcripts per million (TPMs) were obtained from the Genotype-Tissue Expression (GTEx) project V8 [33].
- RNA-Seq in postnatal day 0 (P0) mouse hair cells and non-hair cells from the cochlea [34] from the gene Expression Analysis Resource (gEAR) portal (https://umgear.org, accessed on 17 November 2023). The reads per kilobase of transcript (RPKMs) from the orthologous genes were extracted.
- RNA expression by microarray in P0 mouse spiral ganglion neurons (SGNs) [35] from the Shared Harvard Inner-Ear Laboratory Database (SHIELD, https://shield.hms.harvard.edu, accessed on 17 November 2023). The expression levels were obtained from the orthologs of the candidate genes.
3. Results
3.1. Candidate Variants
3.2. Structural Variants Shared between Individuals with MD and Severe Tinnitus
3.3. Candidate Variants in the Reference Populations
3.4. Expression of Genes with Candidate Variants in the Brain and Inner Ear
4. Discussion
5. Conclusions
- Rare SVs were found in several unrelated individuals with MD and severe tinnitus;
- Three deletions in the ERBB3 gene were identified in two individuals with severe tinnitus;
- A large duplication overlapping the candidate genes AP4M1, COPS6, MCM7, and TAF6 and the non-coding genes MIR106B, MIR25, and MIR93 was carried by two MD patients with severe tinnitus;
- These findings support the role of large SVs in shaping the genetic architecture of severe tinnitus in MD and define new candidate genes associated with this phenotype.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Henton, A.; Tzounopoulos, T. What’s the Buzz? The Neuroscience and the Treatment of Tinnitus. Physiol. Rev. 2021, 101, 1609–1632. [Google Scholar] [CrossRef] [PubMed]
- Online Mendelian Inheritance in Man, OMIM®. Johns Hopkins University, Baltimore, MD. MIM Number: 156000:04/26/2022. Available online: https://omim.org/ (accessed on 13 November 2023).
- Perez-Carpena, P.; Lopez-Escamez, J.A. Current Understanding and Clinical Management of Meniere’s Disease: A Systematic Review. Semin. Neurol. 2020, 40, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Maas, I.L.; Brüggemann, P.; Requena, T.; Bulla, J.; Edvall, N.K.; Hjelmborg, J.V.B.; Szczepek, A.J.; Canlon, B.; Mazurek, B.; Lopez-Escamez, J.A.; et al. Genetic Susceptibility to Bilateral Tinnitus in a Swedish Twin Cohort. Genet. Med. 2017, 19, 1007–1012. [Google Scholar] [CrossRef] [PubMed]
- Cederroth, C.R.; PirouziFard, M.; Trpchevska, N.; Idrizbegovic, E.; Canlon, B.; Sundquist, J.; Sundquist, K.; Zöller, B. Association of Genetic vs. Environmental Factors in Swedish Adoptees with Clinically Significant Tinnitus. JAMA Otolaryngol. Head Neck Surg. 2019, 145, 222–229. [Google Scholar] [CrossRef]
- Trpchevska, N.; Bulla, J.; Prada Hellberg, M.; Edvall, N.K.; Lazar, A.; Mehraei, G.; Uhlen, I.; Schlee, W.; Canlon, B.; Gallus, S.; et al. Sex-Dependent Aggregation of Tinnitus in Swedish Families. J. Clin. Med. 2020, 9, 3812. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Gerstein, M. Unified Views on Variant Impact across Many Diseases. Trends Genet. 2023, 39, 442–450. [Google Scholar] [CrossRef] [PubMed]
- De Ridder, D.; Schlee, W.; Vanneste, S.; Londero, A.; Weisz, N.; Kleinjung, T.; Shekhawat, G.S.; Elgoyhen, A.B.; Song, J.-J.; Andersson, G.; et al. Tinnitus and Tinnitus Disorder: Theoretical and Operational Definitions (an International Multidisciplinary Proposal). Prog. Brain Res. 2021, 260, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Amanat, S.; Gallego-Martinez, A.; Sollini, J.; Perez-Carpena, P.; Espinosa-Sanchez, J.M.; Aran, I.; Soto-Varela, A.; Batuecas-Caletrio, A.; Canlon, B.; May, P.; et al. Burden of Rare Variants in Synaptic Genes in Patients with Severe Tinnitus: An Exome Based Extreme Phenotype Study. EBioMedicine 2021, 66, 103309. [Google Scholar] [CrossRef]
- Gallego-Martinez, A.; Escalera-Balsera, A.; Trpchevska, N.; Robles-Bolivar, P.; Roman-Naranjo, P.; Frejo, L.; Perez-Carpena, P.; Bulla, J.; Gallus, S.; Canlon, B.; et al. Using Coding and Non-Coding Rare Variants to Target Candidate Genes in Patients with Severe Tinnitus. NPJ Genom. Med. 2022, 7, 70. [Google Scholar] [CrossRef]
- Mohan, A.; Leong, S.L.; De Ridder, D.; Vanneste, S. Symptom Dimensions to Address Heterogeneity in Tinnitus. Neurosci. Biobehav. Rev. 2022, 134, 104542. [Google Scholar] [CrossRef]
- Lopez-Escamez, J.A.; Carey, J.; Chung, W.-H.; Goebel, J.A.; Magnusson, M.; Mandalà, M.; Newman-Toker, D.E.; Strupp, M.; Suzuki, M.; Trabalzini, F.; et al. Diagnostic Criteria for Menière’s Disease. J. Vestib. Res. 2015, 25, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag: New York, NY, USA, 2016. [Google Scholar]
- Wickham, H.; Girlich, M. Tidyr: Tidy Messy Data; 2022. Available online: https://tidyr.tidyverse.org (accessed on 11 November 2023).
- Kassambara, A. Ggpubr: “ggplot2” Based Publication Ready Plots; 2020. Available online: https://cran.r-project.org/web/packages/ggpubr/index.html (accessed on 11 November 2023).
- Wickham, H.; Francois, R.; Henry, L.; Müller, K. Dplyr: A Grammar of Data Manipulation; 2022. Available online: https://dplyr.tidyverse.org (accessed on 11 November 2023).
- Wickham, H.; Seidel, D. Scales: Scale Functions for Visualization; 2022. Available online: https://scales.r-lib.org (accessed on 11 November 2023).
- World Medical Association. World Medical Association Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects. JAMA 2013, 310, 2191–2194. [Google Scholar] [CrossRef] [PubMed]
- Szczepek, A.J.; Frejo, L.; Vona, B.; Trpchevska, N.; Cederroth, C.R.; Caria, H.; Lopez-Escamez, J.A. Recommendations on Collecting and Storing Samples for Genetic Studies in Hearing and Tinnitus Research. Ear Hear. 2019, 40, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.; Juhos, S.; Larsson, M.; Olason, P.I.; Martin, M.; Eisfeldt, J.; DiLorenzo, S.; Sandgren, J.; Díaz De Ståhl, T.; Ewels, P.; et al. Sarek: A Portable Workflow for Whole-Genome Sequencing Analysis of Germline and Somatic Variants. F1000Res 2020, 9, 63. [Google Scholar] [CrossRef]
- Ewels, P.A.; Peltzer, A.; Fillinger, S.; Alneberg, J.; Patel, H.; Wilm, A.; Garcia, M.U.; Tommaso, P.D.; Nahnsen, S. Nf-Core: Community Curated Bioinformatics Pipelines. Nat. Biotechnol. 2020, 38, 276–278. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; O’Connor, B.D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra, 1st ed.; O’Reilly Media: Sebastopol, CA, USA, 2020; ISBN 978-1-4919-7519-0. [Google Scholar]
- Talevich, E.; Shain, A.H.; Botton, T.; Bastian, B.C. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput. Biol. 2016, 12, e1004873. [Google Scholar] [CrossRef]
- Chen, X.; Schulz-Trieglaff, O.; Shaw, R.; Barnes, B.; Schlesinger, F.; Källberg, M.; Cox, A.J.; Kruglyak, S.; Saunders, C.T. Manta: Rapid Detection of Structural Variants and Indels for Germline and Cancer Sequencing Applications. Bioinformatics 2016, 32, 1220–1222. [Google Scholar] [CrossRef]
- Eisfeldt, J.; Vezzi, F.; Olason, P.; Nilsson, D.; Lindstrand, A. TIDDIT, an Efficient and Comprehensive Structural Variant Caller for Massive Parallel Sequencing Data. F1000Res 2017, 6, 664. [Google Scholar] [CrossRef]
- Li, H. A Statistical Framework for SNP Calling, Mutation Discovery, Association Mapping and Population Genetical Parameter Estimation from Sequencing Data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Geoffroy, V.; Lamouche, J.-B.; Guignard, T.; Nicaise, S.; Kress, A.; Scheidecker, S.; Le Béchec, A.; Muller, J. The AnnotSV Webserver in 2023: Updated Visualization and Ranking. Nucleic Acids Res. 2023, 51, W39–W45. [Google Scholar] [CrossRef] [PubMed]
- Eisfeldt, J. SVDB 2023. Available online: https://github.com/J35P312/SVDB (accessed on 13 November 2023).
- Shyr, C.; Tarailo-Graovac, M.; Gottlieb, M.; Lee, J.J.; van Karnebeek, C.; Wasserman, W.W. FLAGS, Frequently Mutated Genes in Public Exomes. BMC Med. Genom. 2014, 7, 64. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The Mutational Constraint Spectrum Quantified from Variation in 141,456 Humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- López-López, D.; Roldán, G.; Fernández-Rueda, J.L.; Bostelmann, G.; Carmona, R.; Aquino, V.; Perez-Florido, J.; Ortuño, F.; Pita, G.; Núñez-Torres, R.; et al. A Crowdsourcing Database for the Copy-Number Variation of the Spanish Population. Hum. Genom. 2023, 17, 20. [Google Scholar] [CrossRef] [PubMed]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Cai, T.; Jen, H.-I.; Kang, H.; Klisch, T.J.; Zoghbi, H.Y.; Groves, A.K. Characterization of the Transcriptome of Nascent Hair Cells and Identification of Direct Targets of the Atoh1 Transcription Factor. J. Neurosci. 2015, 35, 5870–5883. [Google Scholar] [CrossRef]
- Lu, C.C.; Appler, J.M.; Houseman, E.A.; Goodrich, L.V. Developmental Profiling of Spiral Ganglion Neurons Reveals Insights into Auditory Circuit Assembly. J. Neurosci. 2011, 31, 10903–10918. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Biswas, R.; Genitsaridi, E.; Trpchevska, N.; Lugo, A.; Schlee, W.; Cederroth, C.R.; Gallus, S.; Hall, D.A. Low Evidence for Tinnitus Risk Factors: A Systematic Review and Meta-Analysis. J. Assoc. Res. Otolaryngol. 2023, 24, 81–94. [Google Scholar] [CrossRef]
- Coey, J.G.; De Jesus, O. Hyperacusis. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Aazh, H.; Taylor, L.; Danesh, A.A.; Moore, B.C.J. The Effectiveness of Unguided Internet-Based Cognitive Behavioral Therapy for Tinnitus for Patients with Tinnitus Alone or Combined with Hyperacusis and/or Misophonia: A Preliminary Analysis. J. Am. Acad. Audiol. 2023, Online ahead of print. [Google Scholar] [CrossRef]
- Herraiz, C.; Tapia, M.C.; Plaza, G. Tinnitus and Ménière’s Disease: Characteristics and Prognosis in a Tinnitus Clinic Sample. Eur. Arch. Otorhinolaryngol. 2006, 263, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Ueberfuhr, M.A.; Wiegrebe, L.; Krause, E.; Gürkov, R.; Drexl, M. Tinnitus in Normal-Hearing Participants after Exposure to Intense Low-Frequency Sound and in Ménière’s Disease Patients. Front. Neurol. 2017, 7, 239. [Google Scholar] [CrossRef] [PubMed]
- Gontika, M.P.; Anagnostouli, M.C. Anti-Myelin Oligodendrocyte Glycoprotein and Human Leukocyte Antigens as Markers in Pediatric and Adolescent Multiple Sclerosis: On Diagnosis, Clinical Phenotypes, and Therapeutic Responses. Mult. Scler. Int. 2018, 2018, 8487471. [Google Scholar] [CrossRef] [PubMed]
- Frejo, L.; Martin-Sanz, E.; Teggi, R.; Trinidad, G.; Soto-Varela, A.; Santos-Perez, S.; Manrique, R.; Perez, N.; Aran, I.; Almeida-Branco, M.S.; et al. Extended Phenotype and Clinical Subgroups in Unilateral Meniere Disease: A Cross-Sectional Study with Cluster Analysis. Clin. Otolaryngol. 2017, 42, 1172–1180. [Google Scholar] [CrossRef] [PubMed]
- Moleon, M.-D.-C.; Martinez-Gomez, E.; Flook, M.; Peralta-Leal, A.; Gallego, J.A.; Sanchez-Gomez, H.; Montilla-Ibañez, M.A.; Dominguez-Durán, E.; Soto-Varela, A.; Aran, I.; et al. Clinical and Cytokine Profile in Patients with Early and Late Onset Meniere Disease. J. Clin. Med. 2021, 10, 4052. [Google Scholar] [CrossRef] [PubMed]
- Raabe, T.D.; Deadwyler, G.; Varga, J.W.; Devries, G.H. Localization of Neuregulin Isoforms and erbB Receptors in Myelinating Glial Cells. Glia 2004, 45, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Torii, T.; Miyamoto, Y.; Takada, S.; Tsumura, H.; Arai, M.; Nakamura, K.; Ohbuchi, K.; Yamamoto, M.; Tanoue, A.; Yamauchi, J. In Vivo Knockdown of ErbB3 in Mice Inhibits Schwann Cell Precursor Migration. Biochem. Biophys. Res. Commun. 2014, 452, 782–788. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Gene Ontology Consortium; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology Knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef]
- Smith, C.L.; Eppig, J.T. The Mammalian Phenotype Ontology: Enabling Robust Annotation and Comparative Analysis. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009, 1, 390–399. [Google Scholar] [CrossRef]
- Fallon, M.; Tadi, P. Histology, Schwann Cells; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Fetoni, A.R.; Troiani, D.; Petrosini, L.; Paludetti, G. Cochlear Injury and Adaptive Plasticity of the Auditory Cortex. Front. Aging Neurosci. 2015, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Møller, A.R.; Langguth, B.; DeRidder, D.; Kleinjung, T. Textbook of Tinnitus; Springer Science & Business Media: New York, NY, USA, 2010; ISBN 978-1-60761-145-5. [Google Scholar]
- Henry, J.A.; Roberts, L.E.; Caspary, D.M.; Theodoroff, S.M.; Salvi, R.J. Underlying Mechanisms of Tinnitus: Review and Clinical Implications. J. Am. Acad. Audiol. 2014, 25, 5–126. [Google Scholar] [CrossRef] [PubMed]
- Bieniossek, C.; Papai, G.; Schaffitzel, C.; Garzoni, F.; Chaillet, M.; Scheer, E.; Papadopoulos, P.; Tora, L.; Schultz, P.; Berger, I. The Architecture of Human General Transcription Factor TFIID Core Complex. Nature 2013, 493, 699–702. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Pehlivan, D.; Karaca, E.; Patel, N.; Charng, W.-L.; Gambin, T.; Gonzaga-Jauregui, C.; Sutton, V.R.; Yesil, G.; Bozdogan, S.T.; et al. Global Transcriptional Disturbances Underlie Cornelia de Lange Syndrome and Related Phenotypes. J. Clin. Investig. 2015, 125, 636–651. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-Z.; Feng, J.-H.; Sun, L.-P.; Ma, H.-W.; Wang, W.-Q.; Li, J.-Y. Novel Compound Heterozygous Variants in the TAF6 Gene in a Patient with Alazami-Yuan Syndrome: A Case Report. World J. Clin. Cases 2022, 10, 1889–1895. [Google Scholar] [CrossRef]
- Akbar, W.; Ullah, A.; Haider, N.; Suleman, S.; Khan, F.U.; Shah, A.A.; Sikandar, M.A.; Basit, S.; Ahmad, W. Identification of Novel Homozygous Variants in FOXE3 and AP4M1 Underlying Congenital Syndromic Anophthalmia and Microphthalmia. J. Gene Med. 2023, e3601. [Google Scholar] [CrossRef]
- Verkerk, A.J.M.H.; Schot, R.; Dumee, B.; Schellekens, K.; Swagemakers, S.; Bertoli-Avella, A.M.; Lequin, M.H.; Dudink, J.; Govaert, P.; van Zwol, A.L.; et al. Mutation in the AP4M1 Gene Provides a Model for Neuroaxonal Injury in Cerebral Palsy. Am. J. Hum. Genet. 2009, 85, 40–52. [Google Scholar] [CrossRef]
- Gou, K.; Liu, J.; Feng, X.; Li, H.; Yuan, Y.; Xing, C. Expression of Minichromosome Maintenance Proteins (MCM) and Cancer Prognosis: A Meta-Analysis. J. Cancer 2018, 9, 1518–1526. [Google Scholar] [CrossRef]
- Snyder, M.; Huang, X.-Y.; Zhang, J.J. The Minichromosome Maintenance Proteins 2-7 (MCM2-7) Are Necessary for RNA Polymerase II (Pol II)-Mediated Transcription. J. Biol. Chem. 2009, 284, 13466–13472. [Google Scholar] [CrossRef]
- Huang, H.-Y.; Lin, Y.-C.-D.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. miRTarBase Update 2022: An Informative Resource for Experimentally Validated miRNA-Target Interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef]
- Bhatt, I.S.; Wilson, N.; Dias, R.; Torkamani, A. A Genome-Wide Association Study of Tinnitus Reveals Shared Genetic Links to Neuropsychiatric Disorders. Sci. Rep. 2022, 12, 22511. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Niu, Y.; Ping, J.; Wang, Y.; Yang, C.; Li, Y.; Zhou, G. Genome-Wide Association Study Identifies New Loci Associated with Noise-Induced Tinnitus in Chinese Populations. BMC Genom. Data 2021, 22, 31. [Google Scholar] [CrossRef] [PubMed]
- Wells, H.R.R.; Abidin, F.N.Z.; Freidin, M.B.; Williams, F.M.K.; Dawson, S.J. Genome-Wide Association Study Suggests That Variation at the RCOR1 Locus Is Associated with Tinnitus in UK Biobank. Sci. Rep. 2021, 11, 6470. [Google Scholar] [CrossRef] [PubMed]
- Urbanek, M.E.; Zuo, J. Genetic Predisposition to Tinnitus in the UK Biobank Population. Sci. Rep. 2021, 11, 18150. [Google Scholar] [CrossRef] [PubMed]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and Limitations of Genome-Wide Association Studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]



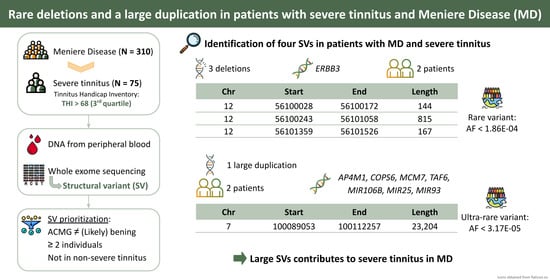
| Chr | Start | End | Length (bp) | SV Type | Individuals | Gene Symbol | ACMG |
|---|---|---|---|---|---|---|---|
| 12 | 56,100,028 | 56,100,172 | 144 | Deletion | I4-40, I4-41 | ERBB3 | US |
| 12 | 56,100,243 | 56,101,058 | 815 | Deletion | I4-40, I4-41 | ERBB3 | US |
| 12 | 56,101,359 | 56,101,526 | 167 | Deletion | I4-40, I4-41 | ERBB3 | US |
| 7 | 100,089,053 | 100,112,257 | 23,204 | Duplication | I4-28, I4-37 | AP4M1, COPS6, MCM7, MIR106B, MIR25, MIR93, TAF6 | US |
| Chr | Start | End | Length (bp) | SV Type | gnomAD NFE | gnomAD | ||
|---|---|---|---|---|---|---|---|---|
| Frequency | Number of Individuals | Frequency | Number of Individuals | |||||
| 12 | 56,100,029 | 56,100,167 | 138 | Deletion | 1.02 × 104 | 6 | 8.74 × 10−5 | 11 |
| 12 | 56,099,811 | 56,101,058 | 1247 | Deletion | 1.69 × 105 | 1 | 2.38 × 10−5 | 3 |
| 12 | 56,100,244 | 56,101,058 | 814 | Deletion | 1.36 × 104 | 8 | 1.27 × 10−4 | 15 |
| 12 | 56,101,363 | 56,101,526 | 163 | Deletion | 1.86 × 104 | 11 | 1.35 × 10−4 | 14 |
| 7 | 100,076,358 | 100,093,492 | 17,134 | Duplication | 3.39 × 105 | 2 | 3.17 × 10−5 | 4 |
| 7 | 100,056,545 | 100,214,180 | 157,635 | Duplication | 0 | 0 | 7.93 × 10−6 | 1 |
| 7 | 100,095,604 | 100,095,982 | 378 | Duplication | 0 | 0 | 7.93 × 10−6 | 1 |
| Gene Symbol | LOEUF | pLI |
|---|---|---|
| ERBB3 | 0.685 | 0 |
| AP4M1 | 1.107 | 0 |
| COPS6 | 0.374 | 1 |
| MCM7 | 1.307 | 0 |
| TAF6 | 0.657 | 0.55 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Escalera-Balsera, A.; Parra-Perez, A.M.; Gallego-Martinez, A.; Frejo, L.; Martin-Lagos, J.; Rivero de Jesus, V.; Pérez-Vázquez, P.; Perez-Carpena, P.; Lopez-Escamez, J.A. Rare Deletions or Large Duplications Contribute to Genetic Variation in Patients with Severe Tinnitus and Meniere Disease. Genes 2024, 15, 22. https://doi.org/10.3390/genes15010022
Escalera-Balsera A, Parra-Perez AM, Gallego-Martinez A, Frejo L, Martin-Lagos J, Rivero de Jesus V, Pérez-Vázquez P, Perez-Carpena P, Lopez-Escamez JA. Rare Deletions or Large Duplications Contribute to Genetic Variation in Patients with Severe Tinnitus and Meniere Disease. Genes. 2024; 15(1):22. https://doi.org/10.3390/genes15010022
Chicago/Turabian StyleEscalera-Balsera, Alba, Alberto M. Parra-Perez, Alvaro Gallego-Martinez, Lidia Frejo, Juan Martin-Lagos, Victoria Rivero de Jesus, Paz Pérez-Vázquez, Patricia Perez-Carpena, and Jose A. Lopez-Escamez. 2024. "Rare Deletions or Large Duplications Contribute to Genetic Variation in Patients with Severe Tinnitus and Meniere Disease" Genes 15, no. 1: 22. https://doi.org/10.3390/genes15010022