Genetic Diversity in the mtDNA of Physarum polycephalum
Abstract
:1. Introduction
Mitochondrial DNA Evolution
2. The Mitochondrial DNA of P. polycephalum
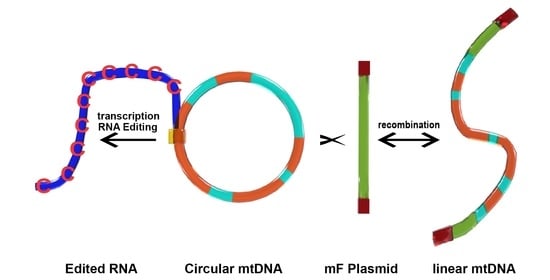
2.1. Mitochondrial Insertional Cotranscriptional RNA Editing in the Myxomycetes (MICOTREM)
Evolution of RNA Editing Site Location
2.2. Unidentified, Untranscribed but Significant Open Reading Frames in the mtDNA of P. polycephalum
2.3. Mitochondrial Open Reading Frames Derived from the mF Mitochondrial Plasmid
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gray, M.W.; Burger, G.; Lang, B.F. Mitochondrial Evolution. Science 1999, 283, 1476–1482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorsness, P.E.; Hanekamp, T. Mitochondria: Origin; John Wiley & Sons, Ltd.: New York, NY, USA, 2001. [Google Scholar] [CrossRef]
- Kolesnikov, A.A.; Gerasimov, E.S. Diversity of mitochondrial genome organization. Biochemistry 2012, 77, 1424–1435. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R.; Keeling, P.J. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc. Natl. Acad. Sci. USA 2015, 112, 10177–10184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavrov, D.V.; Pett, W. Animal mitochondrial DNA as we do not know it: Mt-genome organization and evolution in nonbilaterian lineages. Genome Biol. Evol. 2016, 8, 2896–2913. [Google Scholar] [CrossRef] [PubMed]
- Gualberto, J.M.; Mileshina, D.; Wallet, C.; Niazi, A.K.; Weber-Lotfi, F.; Dietrich, A. The plant mitochondrial genome: Dynamics and maintenance. Biochimie 2014, 100, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Zikova, A.; Hampl, V.; Paris, Z.; Tyc, J.; Lukes, J. Aerobic mitochondria of parasitic protists: Diverse genomes and complex functions. Mol. Biochem. Parisitol. 2016, 209, 45–57. [Google Scholar]
- Takano, H.; Abe, T.; Sakurai, R.; Moriyama, Y.; Miyazawa, Y.; Nozaki, H.; Kawano, S.; Sasaki, N.; Kuroiwa, T. The complete DNA sequence of the mitochondrial DNA of P. polycephalum. Mol. Gen. Genet. 2001, 264, 539–545. [Google Scholar] [CrossRef]
- Miller, D.; Padmanabhan, R.; Sarcar, S.N. Genomics and gene expression in myxomycetes. In Myxomycetes: Biology, Systematics, Biogeography and Ecology, 2nd ed.; Rojas, C., Stephenson, S.L., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 153–193. [Google Scholar]
- Jones, E.P.; Mahendran, R.; Spottswood, M.R.; Yang, Y.-C.; Miller, D.L. Mitochondrial DNA of Physarum: Physical Mapping, Cloning, and Transcription Mapping. Curr. Genet. 1990, 17, 331–337. [Google Scholar] [CrossRef]
- Bundschuh, R.; Antmuller, J.; Becker, C.; Nurnburg, P.; Gott, J.M. Complete characterization of the edited transcriptome of the mitochondrion of Physarum polycephalum using deep sequencing of RNA. Nucleic Acids Res. 2011, 39, 6044–6055. [Google Scholar] [CrossRef] [Green Version]
- Houtz, J.; Cremona, N.; Gott, J.M. Editing of mitochondrial RNAs in P. polycephalum. In RNA Metabolism in Mitochondria; Cruz-Reyes, J., Gray, M.W., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 199–222. [Google Scholar]
- Mahendran, R.; Spottswood, M.S.; Miller, D.L. RNA editing by cytidine insertion in mitochondria of P. polycephalum. Nature 1991, 349, 434–438. [Google Scholar] [CrossRef]
- Miller, D.L.; Mahendran, R.; Spottswood, M.S.; Costandy, H.; Ling, M.L.; Yang, N. Insertional editing in mitochondria of Physarum. Semin. Cell Biol. 1993, 4, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.L.; Mahendran, R.; Spottswood, M.S.; Ling, M.L.; Wang, S.; Yang, N.; Costandy, H. RNA editing in mitochondria of P. polycephalum. In RNA Editing: The Alteration of Protein Coding Sequences of RNA; Benne, R., Ed.; Ellis Horwood: Chichester, UK, 1993; pp. 87–103. [Google Scholar]
- Miller, D.L.; Mahendran, R.; Spottswood, M.S.; Ling, M.L.; Wang, S.; Yang, N.; Costandy, H. RNA editing in mitochondria of Physarum polycephalum. In Plant Mitochondria; Brennicke, A., Kuck, U., Eds.; VHC: Weinheim, Germany, 1993; pp. 53–62. [Google Scholar]
- Gott, J.M.; Visomirski, L.M.; Hunter, J.L. Substitutional and insertional RNA editing of the cytochrome c oxidase subunit I mRNA of P. polycephalum. J. Biol. Chem. 1993, 268, 25483–25486. [Google Scholar] [CrossRef] [PubMed]
- Mahendran, R.; Spottswood, M.S.; Ghate, A.; Ling, M.L.; Jeng, K.; Miller, D.L. Editing of the mitochondrial small subunit rRNA in P. polycehalum. EMBO J. 1994, 13, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Antes, T.; Costandy, H.; Mahendran, R.; Spottswood, M.; Miller, D. Insertional editing of tRNAs of P. polycephalum and Didymium nigripes. Mol. Cell Biol. 1998, 18, 7521–7527. [Google Scholar] [CrossRef] [Green Version]
- Krishnan, U.; Barsamian, A.; Miller, D.L. Evolution of RNA Editing Sites in the Mitochondrial Small Subunit rRNA of the Myxomycetes. Methods Enzymol. 2007, 424, 197–220. [Google Scholar]
- Cheng, Y.W.; Visomirski-Robic, L.M.; Gott, J.M. Non-templated addition of nucleotides to the 3′ end of nascent RNA during RNA editing in Physarum. EMBO J. 2001, 20, 1405–1414. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.L.; Miller, D.L. Non-DNA-templated addition of nucleotides to the 3′ end of RNAs by the mitochondrial RNA Polymerase of P. polycephalum. Mol. Cell Biol. 2008, 28, 5795–5802. [Google Scholar] [CrossRef] [Green Version]
- Cruz-Reyes, J.; Mooers, B.H.M.; Kumar, V.; Doharey, P.K.; Meehan, J.; Chapparo, L. Control Mechanisms of the Holo-Editosome in Trypanosomes. In RNA Metabolism in Mitochondria; Cruz-Reyes, J., Gray, M.W., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 125–144. [Google Scholar]
- Miller, M.L.; Antes, T.J.; Qian, F.; Miller, D.L. Identification of a putative mitochondrial RNA polymerase from P. polycephalum: Characterization, expression, purification, and transcription in vitro. Curr. Genet. 2006, 49, 259–271. [Google Scholar] [CrossRef]
- Rhee, A.C.; Somerlot, B.H.; Parmi, N.; Gott, J.M. Distinct roles for sequences upstream of and downstream from Physarum editing sites. RNA 2009, 15, 1753–1765. [Google Scholar] [CrossRef] [Green Version]
- Sarcar, S.N.; Miller, D.L. A specific, promoter-independent activity of T7 RNA polymerase suggests a general model for DNA/RNA editing in single subunit RNA polymerases. Nat. Sci. Rep. 2018, 8, 13885. [Google Scholar] [CrossRef] [Green Version]
- Landweber, L.F. The evolution of RNA editing in kinetoplastid protozoa. Biosystems 1992, 28, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Maslov, D.A.; Avila, H.A.; Lake, J.A.; Simpson, L. Evolution of RNA editing in kinetoplastid protozoa. Nature 1994, 368, 345–348. [Google Scholar] [CrossRef] [PubMed]
- Simpson, L.; Maslov, D.A. Ancient origin of RNA editing in kinetoplastid protozoa. Curr. Opin. Genet. Dev. 1994, 4, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Skaguchi, K. Invertrons, a class of structurally and functionally related genetic elements that includes linear DNA plasmids, Transposable Elements, and genomes of adeno-type viruses. Microbiol. Rev. 1990, 54, 66–74. [Google Scholar] [CrossRef]
- Takano, H.; Kawano, S.; Kuriowa, T. Constitutive homologous recombination between mitochondrial DNA and a linear mitochondrial plasmid in P. polycephalum. Curr. Genet. 1992, 22, 221–227. [Google Scholar] [CrossRef]
- Nakagawa, C.C.; Jones, E.P.; Miller, D.L. Mitochondrial DNA rearrangements associated with mF plasmid integration and plasmodial longevity in P. polycephalum. Curr. Genet. 1998, 33, 178–187. [Google Scholar] [CrossRef]
- Nomura, H.; Moriyama, Y.; Kawano, S. Rearrangements in the Physarum polycephalum mitochondrial genome associated with a transition from linear mF-mtDNA recombinants to circular molecules. Curr. Genet. 2005, 47, 100–110. [Google Scholar] [CrossRef]
- Takano, H.; Mori, K.; Kawano, S.; Kuroiwa, T. Rearrangements of mitochondrial DNA and the mitochondrial fusion-promoting plasmid (mF) are associated with defective mitochondrial fusion in P. polycephalum. Curr. Genet. 1996, 29, 257–264. [Google Scholar] [CrossRef]
- Takano, H.; Kawano, S.; Kuriowa, T. Complex terminal structure of a linear mitochondrial plasmid from P. polycephalum: Three terminal inverted repeats and an ORF encoding DNA polymerase. Curr. Genet. 1994, 25, 252–257. [Google Scholar] [CrossRef]
- Kawano, S.; Takano, H.; Mori, K.; Kuriowa, T. A mitochondrial plasmid that promotes mitochondrial fusion in P. polycephalum. Protoplasma 1991, 160, 167–169. [Google Scholar] [CrossRef]
- Meijer, W.J.J.; Horcajadas, J.A.; Salas, M. Phi29 Family of Phages. Microbiol. Mol. Biol. Rev. 2001, 65, 261–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Symbol | Gene Name | Gene Length (bp) | ClassicalMt Gene | RNA Editing | Transcription Direction | URF Feature |
| Circular Mitochondrial DNA Genes | ||||||
| t2 | tRNAE gene | 69 | C | E | CCW | |
| tRNAM1 gene | 69 | C | E | CCW | ||
| LSU | Large subunit rRNA gene | 2718 | C | E | CW | |
| SSU | Small subunit rRNA gene | 1814 | C | E | CW | |
| 5S | 5S rRNA gene | 96 | C | CW | ||
| t3 | tRNAM2 gene | 72 | C | CW | ||
| tRNAK gene | 72 | C | E | CW | ||
| tRNAP gene | 71 | C | E | CW | ||
| AB | atp B, ATP synthase, subunit B | 293 | C | CW | ||
| AA | atp A, ATP synthase, subunit A | 536 | C | E | CW | |
| C3 | cox 3, cytochrome oxidase subunit 3 | 762 | C | E | CCW | |
| N6 | nad 6, NADH dehydrogenase, subunit 6 | 479 | C | E | CCW | |
| L16 | rpl 16, large subunit ribosomal protein 16 | 525 | C | E | CCW | |
| S3 | rps 3, small subunit ribosomal protein 3 | 1370 | C | E | CCW | |
| N1 | nad 1, NADH dehydrogenase, subunit 1 | 929 | C | E | CW | |
| Cb | cob, cyt b, cytochrome b oxidase | 1136 | C | E | CCW | |
| A6 | atp 9, ATP synthase, subunit 9 | 243 | C | E | CW | |
| J | URF J, Unassigned reading frame J | 1412 | CW | TM | ||
| K | URF K, Unassigned reading frame K | 1062 | CW | TM | ||
| L | URF L, Unassigned reading frame L | 1086 | CW | TM | ||
| M | URF M, Unassigned reading frame M | 2217 | CW | |||
| N | URF N, Unassigned reading frame N | 1200 | CW | TM, Fb | ||
| O | URF O, Unassigned reading frame O | 579 | CW | TM | ||
| P | URF P, Unassigned reading frame P | 1509 | CW | |||
| Q | URF Q, Unassigned reading frame Q | 1167 | CW | TM, Fb | ||
| R | URF R, Unassigned reading frame R | 663 | CW | RS | ||
| S | URF S, Unassigned reading frame S | 1629 | CW | TM | ||
| T | URF T, Unassigned reading frame T | 222 | CW | TM | ||
| U | URF U, Unassigned reading frame U | 1128 | CW | TM | ||
| V | URF V, Unassigned reading frame V | 336 | CW | |||
| W | URF W, Unassigned reading frame W | 393 | CW | |||
| X | URF X, Unassigned reading frame X | 669 | CW | |||
| Y | URF Y, Unassigned reading frame Y | 2172 | CW | RNAP | ||
| N5 | nad 5, NADH dehydrogenase, subunit 5 | 1894 | C | E | CW | |
| NG | nad 11/G, NADH dehydrogenase, subunit 11 | 1017 | C | E | CW | |
| S2 | rps 2, small subunit ribosomal protein 2 | 1356 | C | E | CW | |
| S12 | rps 12, small subunit ribosomal protein 12 | 496 | C | E | CW | |
| S7 | rps 7, small subunit ribosomal protein 7 | 764 | C | E | CW | |
| Z | URF Z, Unassigned reading frame Z | 352 | CW | T | ||
| C1 | cox 1, cytochrome oxidase subunit 1 | 1719 | C | E | CCW | |
| N7 | nad 7, NADH dehydrogenase, subunit 7 | 1065 | C | E | CCW | |
| C2 | cox 2, cytochrome oxidase, subunit 2 | 864 | C | E | CCW | |
| L11 | rpl 11, large subunit ribosomal protein 11 | 877 | C | E | CW | |
| N2 | nad 2, NADH dehydrogenase, subunit 2 | 1407 | C | E | CW | |
| A | URF A, Unassigned reading frame A | 714 | CCW | TM | ||
| B | URF B, Unassigned reading frame B | 1233 | CCW | TM | ||
| C | URF C, Unassigned reading frame C | 222 | CCW | |||
| S16 | rps 16, small subunit ribosomal protein 16 | 525 | C | E | CW | |
| L19 | rpl 19, large subunit ribosomal protein 19 | 553 | C | E | CW | |
| A8 | atp 8, ATP synthase, subunit 8 | 222 | C | E | CW | |
| N4L | nad 4L, NADH dehydrogenase, subunit 4L | 275 | C | E | CW | |
| A6 | atp 6 ATP synthase, subunit 6 | 708 | C | E | CW | |
| N4 | nad 4, NADH dehydrogenase, subunit 4 | 1383 | C | E | CCW | |
| N3 | nad 3, NADH dehydrogenase, subunit 3 | 374 | C | E | CCW | |
| L14 | rpl 14, large subunit ribosomal protein 14 | 354 | C | E | CW | |
| L5 | rpl 5, large subunit ribosomal protein 5 | 485 | C | E | CW | |
| S14 | rps 14, small subunit ribosomal protein 14 | 265 | C | E | CW | |
| S8 | rps 8, small subunit ribosomal protein 8 | 422 | C | E | CW | |
| L6 | rpl 6, large subunit ribosomal protein 6 | 467 | C | E | CW | |
| S13 | rps 13, small subunit ribosomal protein 13 | 539 | C | E | CW | |
| N9 | nad 9, NADH dehydrogenase, subunit 9 | 475 | C | E | CW | |
| S11 | rps 11, small subunit ribosomal protein 11 | 721 | C | E | CW | |
| E1 | ERF 1, edited reading frame 1 | 652 | ? | E | CW | |
| S4 | rps 4, small subunit ribosomal protein 4 | 790 | C | E | CCW | |
| D | URF D, Unassigned reading frame D | 267 | CCW | TM | ||
| E | URF E, Unassigned reading frame E | 612 | CCW | TM | ||
| F | URF F, Unassigned reading frame F | 441 | CCW | |||
| G | URF G, Unassigned reading frame G | 738 | CW | |||
| H | URF H, Unassigned reading frame H | 597 | CW | TM | ||
| I | URF I, Unassigned reading frame I | 390 | CCW | TM | ||
| mF Plasmid Genes | ||||||
| 1 | URF 1, mF unassigned Reading frame 1 | 696 | LTR | |||
| 2 | URF 2, mF unassigned Reading frame 2 | 492 | LTR | |||
| 3 | URF 3, mF unassigned Reading frame 3 | 1923 | LTR | TM | ||
| 4 | URF 4, mF unassigned Reading frame 4 | 708 | LTR | |||
| 5 | URF 5, mF unassigned Reading frame 5 | 357 | LTR | |||
| 6 | URF 6, mF unassigned Reading frame 6 | 3393 | LTR | RNAP | ||
| 7 | URF 7, mF unassigned Reading frame 7 | 1101 | LTR | RS | ||
| 8 | URF 8, mF unassigned Reading frame 8 | 930 | LTR | TM | ||
| 9 | URF 9, mF unassigned Reading frame 9 | 1644 | LTR | DNAP | ||
| Linear mtDNA Hybrid Genes Produced by Recombination | ||||||
| R7 | URF R7, hybrid unassigned Reading frame R7 | 630 | LTR | |||
| 7R | URF 7R, hybrid unassigned Reading frame 7R | 1131 | LTR | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hammar, F.; Miller, D.L. Genetic Diversity in the mtDNA of Physarum polycephalum. Genes 2023, 14, 628. https://doi.org/10.3390/genes14030628
Hammar F, Miller DL. Genetic Diversity in the mtDNA of Physarum polycephalum. Genes. 2023; 14(3):628. https://doi.org/10.3390/genes14030628
Chicago/Turabian StyleHammar, Freya, and Dennis L. Miller. 2023. "Genetic Diversity in the mtDNA of Physarum polycephalum" Genes 14, no. 3: 628. https://doi.org/10.3390/genes14030628