A CRISPR Screen Identifies the E3 Ubiquitin Ligase Rfwd2 as a Negative Regulator of Glucose Uptake in Brown Adipocytes
Abstract
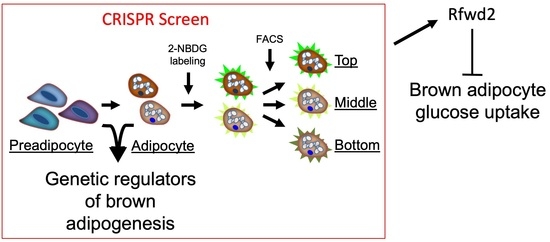
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Viral Transduction
2.3. 2-NBDG Staining and Sorting
2.4. Oil Red O Staining
2.5. 2-DOG Uptake
2.6. PCR
2.7. Immunoblotting
2.8. Proteomics
2.9. Data Analysis and Statistics
3. Results
3.1. Generation of a Mature Brown Adipocyte Cell Library
3.2. Screening 2-NBDG Uptake in Mature Adipocytes
3.3. Validating Putative Genetic Regulators of 2-NBDG Uptake
3.4. Rfwd2 Targeting Decreases Protein Levels and Increases Glucose Uptake
3.5. Identifying Putative Targets of Rfwd2-Mediated Proteolysis in Brown Adipocytes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Townsend, K.L.; Tseng, Y.H. Brown fat fuel utilization and thermogenesis. Trends Endocrinol. Metab. 2014, 25, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.S. Recent insights into the molecular mechanisms of simultaneous fatty acid oxidation and synthesis in brown adipocytes. Front. Endocrinol. 2023, 14, 1106544. [Google Scholar] [CrossRef] [PubMed]
- Cypess, A.M.; Lehman, S.; Williams, G.; Tal, I.; Rodman, D.; Goldfine, A.B.; Kahn, C.R.; Kuo, F.C.; Palmer, E.L.; Tseng, Y.-H.; et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009, 360, 1509–1517. [Google Scholar] [CrossRef]
- Virtanen, K.A.; Lidell, M.E.; Orava, J.; Heglind, M.; Westergren, R.; Niemi, T.; Taittonen, M.; Laine, J.; Savisto, N.-J.; Enerbäck, S.; et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 2009, 360, 1518–1525. [Google Scholar] [CrossRef]
- Van Marken Lichtenbelt, W.D.; Vanhommerig, J.W.; Smulders, N.M.; Drossaerts, J.M.A.F.L.; Kemerink, G.J.; Bouvy, N.D.; Schrauwen, P.; Teule, G.J.J. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 2009, 360, 1500–1508. [Google Scholar] [CrossRef]
- Cypess, A.M.; Chen, Y.-C.; Sze, C.; Wang, K.; English, J.; Chan, O.; Holman, A.R.; Tal, I.; Palmer, M.R.; Kolodny, G.M.; et al. Cold but not sympathomimetics activates human brown adipose tissue in vivo. Proc. Natl. Acad. Sci. USA 2012, 109, 10001–10005. [Google Scholar] [CrossRef] [PubMed]
- Kulterer, O.C.; Niederstaetter, L.; Herz, C.T.; Haug, A.R.; Bileck, A.; Pils, D.; Kautzky-Willer, A.; Gerner, C.; Kiefer, F.W. The Presence of Active Brown Adipose Tissue Determines Cold-Induced Energy Expenditure and Oxylipin Profiles in Humans. J. Clin. Endocrinol. Metab. 2020, 105, 2203–2216. [Google Scholar] [CrossRef]
- Cypess, A.M.; Weiner, L.S.; Roberts-Toler, C.; Elía, E.F.; Kessler, S.H.; Kahn, P.A.; English, J.; Chatman, K.; Trauger, S.A.; Doria, A.; et al. Activation of human brown adipose tissue by a β3-adrenergic receptor agonist. Cell Metab. 2015, 21, 33–38. [Google Scholar] [CrossRef]
- Wibmer, A.G.; Becher, T.; Eljalby, M.; Crane, A.; Andrieu, P.C.; Jiang, C.S.; Vaughan, R.; Schöder, H.; Cohen, P. Brown adipose tissue is associated with healthier body fat distribution and metabolic benefits independent of regional adiposity. Cell Rep. Med. 2021, 2, 100332. [Google Scholar] [CrossRef]
- Becher, T.; Palanisamy, S.; Kramer, D.J.; Eljalby, M.; Marx, S.J.; Wibmer, A.G.; Butler, S.D.; Jiang, C.S.; Vaughan, R.; Schöder, H.; et al. Brown adipose tissue is associated with cardiometabolic health. Nat. Med. 2021, 27, 58–65. [Google Scholar] [CrossRef]
- Lynes, M.D.; Tseng, Y.H. The Thermogenic Circuit: Regulators of Thermogenic Competency and Differentiation. Genes. Dis. 2015, 2, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Calejman, C.M.; Doxsey, W.G.; Fazakerley, D.J.; Guertin, D.A. Integrating adipocyte insulin signaling and metabolism in the multi-omics era. Trends Biochem. Sci. 2022, 47, 531–546. [Google Scholar] [CrossRef]
- Lebensohn, A.M.; Dubey, R.; Neitzel, L.R.; Tacchelly-Benites, O.; Yang, E.; Marceau, C.D.; Davis, E.M.; Patel, B.B.; Bahrami-Nejad, Z.; Travaglini, K.J.; et al. Comparative genetic screens in human cells reveal new regulatory mechanisms in WNT signaling. eLife 2016, 5, e21459. [Google Scholar] [CrossRef] [PubMed]
- Parnas, O.; Jovanovic, M.; Eisenhaure, T.M.; Herbst, R.H.; Dixit, A.; Ye, C.J.; Przybylski, D.; Platt, R.J.; Tirosh, I.; Sanjana, N.E.; et al. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell 2015, 162, 675–686. [Google Scholar] [CrossRef]
- Gulbranson, D.R.; Davis, E.M.; Demmitt, B.A.; Ouyang, Y.; Ye, Y.; Yu, H.; Shen, J. RABIF/MSS4 is a Rab-stabilizing holdase chaperone required for GLUT4 exocytosis. Proc. Natl. Acad. Sci. USA 2017, 114, E8224–E8233. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Fujimoto, M.; Lopes, A.; Wang, Y.X. Twist-1 is a PPARdelta-inducible, negative-feedback regulator of PGC-1alpha in brown fat metabolism. Cell 2009, 137, 73–86. [Google Scholar] [CrossRef]
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.; Orchard, R.; et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [CrossRef]
- Ortega-Molina, A.; Efeyan, A.; Lopez-Guadamillas, E.; Muñoz-Martin, M.; Gómez-López, G.; Cañamero, M.; Mulero, F.; Pastor, J.; Martinez, S.; Romanos, E.; et al. Pten positively regulates brown adipose function, energy expenditure, and longevity. Cell Metab. 2012, 15, 382–394. [Google Scholar] [CrossRef]
- Sanchez-Gurmaches, J.; Hung, C.M.; Sparks, C.A.; Tang, Y.; Li, H.; Guertin, D.A. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metab. 2012, 16, 348–362. [Google Scholar] [CrossRef]
- Yoshida, A.; Kato, J.Y.; Nakamae, I.; Yoneda-Kato, N. COP1 targets C/EBPα for degradation and induces acute myeloid leukemia via Trib1. Blood 2013, 122, 1750–1760. [Google Scholar] [CrossRef]
- Dornan, D.; Wertz, I.; Shimizu, H.; Arnott, D.; Frantz, G.D.; Dowd, P.; O’Rourke, K.; Koeppen, H.; Dixit, V.M. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 2004, 429, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Wertz, I.E.; O’Rourke, K.M.; Zhang, Z.; Dornan, D.; Arnott, D.; Deshaies, R.J.; Dixit, V.M. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 2004, 303, 1371–1374. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Heredia, J.E.; Altarejos, J.Y.; Screaton, R.; Goebel, N.; Niessen, S.; MacLeod, I.X.; Liew, C.W.; Kulkarni, R.N.; Bain, J.; et al. TRB3 links the E3 ubiquitin ligase COP1 to lipid metabolism. Science 2006, 312, 1763–1766. [Google Scholar] [CrossRef]
- Ghosh, M.; Niyogi, S.; Bhattacharyya, M.; Adak, M.; Nayak, D.K.; Chakrabarti, S.; Chakrabarti, P. Ubiquitin Ligase COP1 Controls Hepatic Fat Metabolism by Targeting ATGL for Degradation. Diabetes 2016, 65, 3561–3572. [Google Scholar] [CrossRef]
- Orlicky, D.J.; Schaack, J. Adenovirus transduction of 3T3-L1 cells. J. Lipid Res. 2001, 42, 460–466. [Google Scholar] [CrossRef]
- Carlotti, F.; Bazuine, M.; Kekarainen, T.; Seppen, J.; Pognonec, P.; Maassen, J.; Hoeben, R.C. Lentiviral vectors efficiently transduce quiescent mature 3T3-L1 adipocytes. Mol. Ther. 2004, 9, 209–217. [Google Scholar] [CrossRef]
- Jiang, Z.Y.; Zhou, Q.L.; Coleman, K.A.; Chouinard, M.; Boese, Q.; Czech, M.P. Insulin signaling through Akt/protein kinase B analyzed by small interfering RNA-mediated gene silencing. Proc. Natl. Acad. Sci. USA 2003, 100, 7569–7574. [Google Scholar] [CrossRef]
- Zou, C.; Wang, Y.; Shen, Z. 2-NBDG as a fluorescent indicator for direct glucose uptake measurement. J. Biochem. Biophys. Methods 2005, 64, 207–215. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, R.G.; Wu, L.; Mullani, N. Uptake of a fluorescent deoxyglucose analog (2-NBDG) in tumor cells. Mol. Imaging Biol. 2005, 7, 388–392. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lynes, M.D.; Huang, Q.; Cora, C.; Su, S.-C.; Yi, P.; Tseng, Y.-H. A CRISPR Screen Identifies the E3 Ubiquitin Ligase Rfwd2 as a Negative Regulator of Glucose Uptake in Brown Adipocytes. Genes 2023, 14, 1865. https://doi.org/10.3390/genes14101865
Lynes MD, Huang Q, Cora C, Su S-C, Yi P, Tseng Y-H. A CRISPR Screen Identifies the E3 Ubiquitin Ligase Rfwd2 as a Negative Regulator of Glucose Uptake in Brown Adipocytes. Genes. 2023; 14(10):1865. https://doi.org/10.3390/genes14101865
Chicago/Turabian StyleLynes, Matthew D., Qian Huang, Carolina Cora, Sheng-Chiang Su, Peng Yi, and Yu-Hua Tseng. 2023. "A CRISPR Screen Identifies the E3 Ubiquitin Ligase Rfwd2 as a Negative Regulator of Glucose Uptake in Brown Adipocytes" Genes 14, no. 10: 1865. https://doi.org/10.3390/genes14101865