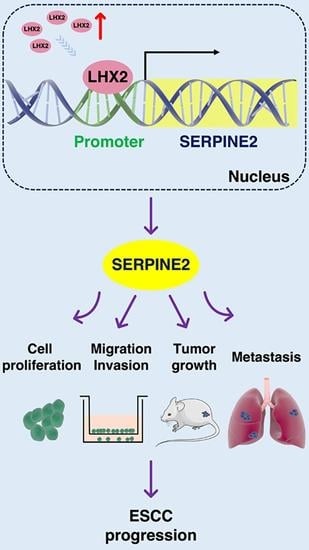
LHX2 Enhances the Malignant Phenotype of Esophageal Squamous Cell Carcinoma by Upregulating the Expression of SERPINE2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture
2.2. Plasmids, Virus Package and Infection
2.3. Western Blotting
2.4. RNA Isolation and Quantitative Real-Time PCR
2.5. CCK-8 Assays and Colony-Formation Ability
2.6. Transwell and Wound-Healing Assays
2.7. Animal Experiments
2.8. RNA-Seq and ChIP Assays
2.9. Statistical Analysis
3. Results
3.1. LHX2 Is Significantly Upregulated in ESCC
3.2. LHX2 Enhances the Proliferation and Tumor Growth of ESCC Cells
3.3. LHX2 Promotes the Migration, Invasion and Metastasis of ESCC Cells
3.4. LHX2 Transcriptionally Regulates the Expression of SERPINE2 in ESCC
3.5. LHX2 Augments the Malignant Phenotype of ESCC by Upregulating SERPINE2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.; Van Lanschot, J.J.B.; Hulshof, M.C.C.M.; van Hagen, P.; van Berge Henegouwen, M.I.; Wijnhoven, B.P.L.; van Laarhoven, H.W.M.; Nieuwenhuijzen, G.A.P.; Hospers, G.A.P.; Bonenkamp, J.J.; et al. Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for oesophageal or junctional cancer (CROSS): Long-term results of a randomised controlled trial. Lancet Oncol. 2015, 16, 1090–1098. [Google Scholar] [CrossRef]
- Chou, S.-J.; Tole, S. Lhx2, an evolutionarily conserved, multifunctional regulator of forebrain development. Brain Res. 2019, 1705, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Singh, D.; Modi, D. LIM Homeodomain (LIM-HD) Genes and Their Co-Regulators in Developing Reproductive System and Disorders of Sex Development. Sex. Dev. 2021, 1–15. [Google Scholar] [CrossRef]
- Singh, N.; Singh, D.; Bhide, A.; Sharma, R.; Sahoo, S.; Jolly, M.K.; Modi, D. Lhx2 in germ cells suppresses endothelial cell migration in the developing ovary. Exp. Cell Res. 2022, 415, 113108. [Google Scholar] [CrossRef]
- Hobert, O.; Westphal, H. Functions of LIM-homeobox genes. Trends Genet. 2000, 16, 75–83. [Google Scholar] [CrossRef]
- Kuzmanov, A.; Hopfer, U.; Marti, P.; Meyer-Schaller, N.; Yilmaz, M.; Christofori, G. LIM-homeobox gene 2 promotes tumor growth and metastasis by inducing autocrine and paracrine PDGF-B signaling. Mol. Oncol. 2014, 8, 401–416. [Google Scholar] [CrossRef]
- Song, H.; Liu, J.; Wu, X.; Zhou, Y.; Chen, X.; Chen, J.; Deng, K.; Mao, C.; Huang, S.; Liu, Z. LHX2 promotes malignancy and inhibits autophagy via mTOR in osteosarcoma and is negatively regulated by miR-129-5p. Aging 2019, 11, 9794–9810. [Google Scholar] [CrossRef]
- Huang, T.; Yang, J.; Liu, B.; Fu, L. A new mouse esophageal cancer cell line (mEC25)-derived pre-clinical syngeneic tumor model for immunotherapy. Cancer Commun. 2020, 40, 316–320. [Google Scholar] [CrossRef]
- Li, X.; Wang, M.; Gong, T.; Lei, X.; Hu, T.; Tian, M.; Ding, F.; Ma, F.; Chen, H.; Liu, Z. A S100A14-CCL2/CXCL5 signaling axis drives breast cancer metastasis. Theranostics 2020, 10, 5687–5703. [Google Scholar] [CrossRef]
- Xie, T.; Du, K.; Liu, W.; Liu, C.; Wang, B.; Tian, Y.; Li, R.; Huang, X.; Lin, J.; Jian, H.; et al. LHX2 facilitates the progression of nasopharyngeal carcinoma via activation of the FGF1/FGFR axis. Br. J. Cancer 2022, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Gou, S.; Xiong, J.; Wu, H.; Wang, C.; Liu, T. Oncogenicity of LHX2 in pancreatic ductal adenocarcinoma. Mol. Biol. Rep. 2014, 41, 8163–8167. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Zhang, L.; Yu, Q.; Zhao, H.; Yang, H.; Wang, Y. Effect of LHX2 gene methylation level and its function on radiotherapy of cervical cancer. Transl. Cancer Res. 2021, 10, 2944–2961. [Google Scholar] [CrossRef] [PubMed]
- Reichenbach, Z.W.; Murray, M.G.; Saxena, R.; Farkas, D.; Karassik, E.G.; Klochkova, A.; Patel, K.; Tice, C.; Hall, T.M.; Gang, J.; et al. Clinical and translational advances in esophageal squamous cell carcinoma. Adv. Cancer Res. 2019, 144, 95–135. [Google Scholar]
- Lin, D.-C.; Wang, M.-R.; Koeffler, H.P. Genomic and Epigenomic Aberrations in Esophageal Squamous Cell Carcinoma and Implications for Patients. Gastroenterology 2018, 154, 374–389. [Google Scholar] [CrossRef]
- Cui, Y.; Chen, H.; Xi, R.; Cui, H.; Zhao, Y.; Xu, E.; Yan, T.; Lu, X.; Huang, F.; Kong, P.; et al. Whole-genome sequencing of 508 patients identifies key molecular features associated with poor prognosis in esophageal squamous cell carcinoma. Cell Res. 2020, 30, 902–913. [Google Scholar] [CrossRef]
- Gao, Y.-B.; Chen, Z.-L.; Li, J.-G.; Hu, X.-D.; Shi, X.-J.; Sun, Z.-M.; Zhang, F.; Zhao, Z.-R.; Li, Z.-T.; Liu, Z.-Y.; et al. Genetic landscape of esophageal squamous cell carcinoma. Nat. Genet. 2014, 46, 1097–1102. [Google Scholar] [CrossRef]
- Tiede, S.; Paus, R. Lhx2--decisive role in epithelial stem cell maintenance, or just the “tip of the iceberg”? Bioessays 2006, 28, 1157–1160. [Google Scholar] [CrossRef]
- Folgueras, A.R.; Guo, X.; Pasolli, H.A.; Stokes, N.; Polak, L.; Zheng, D.; Fuchs, E. Architectural Niche Organization by LHX2 Is Linked to Hair Follicle Stem Cell Function. Cell Stem Cell 2013, 13, 314–327. [Google Scholar] [CrossRef]
- Hsu, L.C.L.; Nam, S.; Cui, Y.; Chang, C.P.; Wang, C.F.; Kuo, H.C.; Touboul, J.D.; Chou, S.J. Lhx2 regulates the timing of β-catenin-dependent cortical neurogenesis. Proc. Natl. Acad. Sci. USA 2015, 112, 12199–12204. [Google Scholar] [CrossRef]
- Mardaryev, A.N.; Meier, N.; Poterlowicz, K.; Sharov, A.A.; Sharova, T.Y.; Ahmed, M.I.; Rapisarda, V.; Lewis, C.; Fessing, M.Y.; Ruenger, T.M.; et al. Lhx2 differentially regulates Sox9, Tcf4 and Lgr5 in hair follicle stem cells to promote epidermal regeneration after injury. Development 2011, 138, 4843–4852. [Google Scholar] [CrossRef] [PubMed]
- Abate-Shen, C. Deregulated homeobox gene expression in cancer: Cause or consequence? Nat. Rev. Cancer 2002, 2, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Sukumar, S. The HOX genes and their roles in oncogenesis. Nat. Rev. Cancer 2010, 10, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Armat, M.; Ramezani, F.; Molavi, O.; Sabzichi, M.; Samadi, N. Six family of homeobox genes and related mechanisms in tumorigenesis protocols. Tumori 2016, 2016, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wan, L.; Xiao, C.; Hu, H.; Wang, L.; Zhao, J.; Lei, Z.; Zhang, H.-T. Inhibition of LHX2 by miR-124 suppresses cellular migration and invasion in non-small cell lung cancer. Oncol. Lett. 2017, 14, 3429–3436. [Google Scholar] [CrossRef]
- Mosca, N.; Khoubai, F.Z.; Fedou, S.; Carrillo-Reixach, J.; Caruso, S.; Del Rio-Alvarez, A.; Dubois, E.; Avignon, C.; Dugot-Senant, N.; Guettier, C.; et al. LIM Homeobox-2 Suppresses Hallmarks of Adult and Pediatric Liver Cancers by Inactivating MAPK/ERK and Wnt/β-Catenin Pathways. Liver Cancer 2022, 11, 126–140. [Google Scholar] [CrossRef]
- Rapa, I.; Votta, A.; Giorcelli, J.; Izzo, S.; Rigutto, A.; Metovic, J.; Napoli, F.; Volante, M. Proposal of a Panel of Genes Identified by miRNA Profiling as Candidate Prognostic Biomarkers in Lung Carcinoids. Neuroendocrinology 2021, 111, 115–122. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, Q.; Zhu, L.; Xie, S.; Tu, L.; Yang, Y.; Wu, K.; Zhao, Y.; Wang, Y.; Xu, Y.; et al. SERPINE2/PN-1 regulates the DNA damage response and radioresistance by activating ATM in lung cancer. Cancer Lett. 2022, 524, 268–283. [Google Scholar] [CrossRef]
- Bergeron, S.; Lemieux, E.; Durand, V.; Cagnol, S.; Carrier, J.C.; Lussier, J.G.; Boucher, M.-J.; Rivard, N. The serine protease inhibitor serpinE2 is a novel target of ERK signaling involved in human colorectal tumorigenesis. Mol. Cancer 2010, 9, 271. [Google Scholar] [CrossRef]
- Wagenblast, E.; Soto, M.; Gutiérrez-Ángel, S.; Hartl, C.A.; Gable, A.L.; Maceli, A.R.; Erard, N.; Williams, A.M.; Kim, S.Y.; Dickopf, S.; et al. A model of breast cancer heterogeneity reveals vascular mimicry as a driver of metastasis. Nature 2015, 520, 358–362. [Google Scholar] [CrossRef]
- Dokuni, R.; Nagano, T.; Jimbo, N.; Sato, H.; Kiriu, T.; Yasuda, Y.; Yamamoto, M.; Tachihara, M.; Kobayashi, K.; Maniwa, Y.; et al. High expression level of serpin peptidase inhibitor clade E member 2 is associated with poor prognosis in lung adenocarcinoma. Respir. Res. 2020, 21, 331. [Google Scholar] [CrossRef] [PubMed]
- Sasahira, T.; Kurihara-Shimomura, M.; Shimomura, H.; Kirita, T. SERPINE2 is an oral cancer-promoting factor that induces angiogenesis and lymphangiogenesis. Int. J. Clin. Oncol. 2021, 26, 1831–1839. [Google Scholar] [CrossRef] [PubMed]
- Chuang, H.W.; Hsia, K.T.; Liao, J.B.; Yeh, C.C.; Kuo, W.T.; Yang, Y.F. SERPINE2 Overexpression Is Associated with Poor Prognosis of Urothelial Carcinoma. Diagnostics 2021, 11, 1928. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Wang, X.; Xu, J.; Lu, L. SerpinE2, a poor biomarker of endometrial cancer, promotes the proliferation and mobility of EC cells. Cancer Biomark. 2017, 19, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.W. Serpine2, a potential novel target for combating melanoma metastasis. Am. J. Transl. Res. 2016, 8, 1985–1997. [Google Scholar]
- Stępień, T.; Brożyna, M.; Kuzdak, K.; Motylewska, E.; Komorowski, J.; Stępień, H.; Ławnicka, H. Elevated Concentrations of SERPINE2/Protease Nexin-1 and Secretory Leukocyte Protease Inhibitor in the Serum of Patients with Papillary Thyroid Cancer. Dis Markers 2017, 2017, 4962137. [Google Scholar] [CrossRef]
- Zhang, J.; Luo, A.; Huang, F.; Gong, T.; Liu, Z. SERPINE2 promotes esophageal squamous cell carcinoma metastasis by activating BMP4. Cancer Lett. 2020, 469, 390–398. [Google Scholar] [CrossRef]





| Sequence | Used for |
|---|---|
| hLHX2-F, 5′-ACGCCAAGGACTTGAAGCAGCT-3′ | qRT-PCR |
| hLHX2-R, 5′-TTTCCTGCCGTAAGAGGTTGCG-3′ | qRT-PCR |
| hβ-actin-F, 5′-AGGCACCAGGGCGTGAT-3′ | qRT-PCR |
| hβ-actin-R, 5′GCCCACATAGGAATCCTTCTGAC-3′ | qRT-PCR |
| mLhx2-F, 5′-GTCATCGACGAGATGGACCG-3′ | qRT-PCR |
| mLhx2-R, 5′-TGAAGCAGGTGAGTTCCGAC-3′ | qRT-PCR |
| mβ-actin-F, 5′-CATTGCTGACAGGATGCAGAAGG-3′ | qRT-PCR |
| mβ-actin-R, 5′-TGCTGGAAGGTGGACAGTGAGG-3′ | qRT-PCR |
| hSERPINE2-F, 5′-AAGAAACGCACTTTCGTGGC-3′ | qRT-PCR |
| hSERPINE2-R, 5′-GTGTGGGATGATGGCAGACA-3′ | qRT-PCR |
| hFGFBP1-F, 5′-CAGGAGGAGGGCATCTCTCT-3′ | qRT-PCR |
| hFGFBP1-R, 5′-TCCGGGCAACTTGTTTCCAA-3′ | qRT-PCR |
| hGBP1-F, 5′-TAGCAGACTTCTGTTCCTACATCT-3′ | qRT-PCR |
| hGBP1-R, 5′-CCACTGCTGATGGCATTGACGT-3′ | qRT-PCR |
| hMAPK6-F, 5′-AGCTTGGGGAGAGAGGACAT-3′ | qRT-PCR |
| hMAPK6-R, 5′-GTGGGATGCCTATGGACTCG-3′ | qRT-PCR |
| SERPINE2-F, 5′-CATTTCTTAACAACTGCATGCCA-3′ | ChIP-qPCR |
| SERPINE2-R, 5′-TCCCTGCATCCATTTCTGTCT-3′ | ChIP-qPCR |
| 5′-GCGCTAAGCTGCAACGAAA-3′ | shLHX2#1 |
| 5′-GCCAGAAGACCAAGCGCAT-3′ | shLHX2#2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Wu, X.; Chen, H.; Liu, Z.; He, H.; Wang, L. LHX2 Enhances the Malignant Phenotype of Esophageal Squamous Cell Carcinoma by Upregulating the Expression of SERPINE2. Genes 2022, 13, 1457. https://doi.org/10.3390/genes13081457
Li X, Wu X, Chen H, Liu Z, He H, Wang L. LHX2 Enhances the Malignant Phenotype of Esophageal Squamous Cell Carcinoma by Upregulating the Expression of SERPINE2. Genes. 2022; 13(8):1457. https://doi.org/10.3390/genes13081457
Chicago/Turabian StyleLi, Xukun, Xueling Wu, Hongyan Chen, Zhihua Liu, Huan He, and Luhua Wang. 2022. "LHX2 Enhances the Malignant Phenotype of Esophageal Squamous Cell Carcinoma by Upregulating the Expression of SERPINE2" Genes 13, no. 8: 1457. https://doi.org/10.3390/genes13081457