Zebularine, a DNA Methylation Inhibitor, Activates Anthocyanin Accumulation in Grapevine Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Anthocyanin, Sugar and Amino Acid Quantification
2.3. Stilbene Quantification
2.4. Nucleic Acid Extraction
2.5. RT-qPCR Analysis
2.6. Methylation Analysis with HPLC and McrBC-PCR
2.7. RNA-Seq Analysis
3. Results
3.1. Zebularine Stimulates Anthocyanin Production
3.1.1. Zebularine Inhibits Cell Growth and Impacts Cell Color
3.1.2. Zebularine Treatment Results in an Increase in the Proportion of Colored Cells
3.1.3. Zebularine Treatments Induce an Increase in Anthocyanin Quantities
3.2. Transcriptome Analysis Suggests Complex Effects of Zebularine on Gene Expression in GT Cells
3.2.1. Genes Involved in the Anthocyanin Pathway Are Differentially Regulated in Light and Dark Grown Cells
3.2.2. Zebularine Treatment Enhances the Whole Flavonoid Biosynthesis Pathway in Illuminated Cells
3.2.3. Zebularine Specifically Induced Genes Associated with Anthocyanin Accumulation in Dark Grown Cells
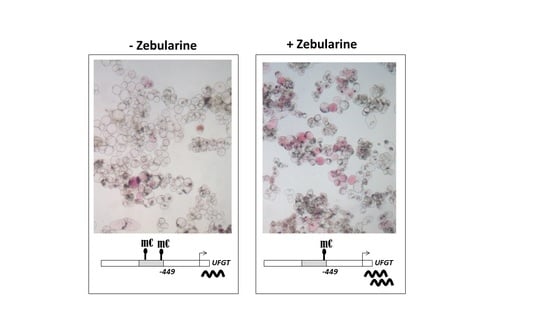
3.3. Anthocyanin Accumulation in Zebularine-Treated Cells Correlates with a Slight Decrease in UFGT Methylation Status
3.4. Zebularine Induces a Stress Response in Grape Cells
Zebularine Slows down Cell Growth and Increases Cell Mortality
4. Discussion
4.1. Zebularine Has a Positive Impact on Anthocyanin Production Irrespective of the Growing Conditions and Mediates Limited Methylation Changes
4.2. Zebularine Induces a Stress-Response and Inhibits the Growth of GT Cell Suspensions
4.3. The Expression Analysis of Anthocyanin-Related Genes Revealed Different Deregulation Patterns in the Dark and in the Light, Suggesting That the Mode of Action of Zebularine Could Be Different in These Two Situations
4.4. Zebularine-Dependent Activation of Anthocyanin Accumulation Was Correlated with an Up-Regulation of UFGT, Together with a Local Decrease in DNA Methylation in UFGT Promoter
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hatier, J.-H.B.; Gould, K.S. Anthocyanin Function in Vegetative Organs. Anthocyanins 2008, 1–19. [Google Scholar]
- Chalker-Scott, L. Environmental Significance of Anthocyanins in Plant Stress Responses. Photochem. Photobiol. 1999, 70, 1–9. [Google Scholar] [CrossRef]
- Ananga, A.; Georgiev, V.; Ochieng, J.; Phills, B.; Tsolov, V. Production of Anthocyanins in Grape Cell Cultures: A Potential Source of Raw Material for Pharmaceutical, Food, and Cosmetic Industries. In The Mediterranean Genetic Code: Grapevine and Olive; Intechopen: London, UK, 2013. [Google Scholar]
- Kuhn, N.; Guan, L.; Dai, Z.W.; Wu, B.-H.; Lauvergeat, V.; Gomès, E.; Li, S.-H.; Godoy, F.; Arce-Johnson, P.; Delrot, S. Berry Ripening: Recently Heard through the Grapevine. J. Exp. Bot. 2013, 65, 4543–4559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin Biosynthesis and Degradation Mechanisms in Vegetables: A Review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef]
- Sparvoli, F.; Martin, C.; Scienza, A.; Gavazzi, G.; Tonelli, C. Cloning and Molecular Analysis of Structural Genes Involved in Flavonoid and Stilbene Biosynthesis in Grape (Vitis Vinifera L.). Plant Mol. Biol. 1994, 24, 743–755. [Google Scholar] [CrossRef]
- Urban, P.; Werck-Reichhart, D.; Teutsch, H.G.; Durst, F.; Regnier, S.; Kazmaier, M.; Pompon, D. Characterization of Recombinant Plant Cinnamate 4-Hydroxylase Produced in Yeast. Kinetic and Spectral Properties of the Major Plant P450 of the Phenylpropanoid Pathway. Eur. J. Biochem. 1994, 222, 843–850. [Google Scholar] [CrossRef]
- Ford, C.M.; Boss, P.K.; Hoj, P.B. Cloning and Characterization of Vitis Vinifera UDP-Glucose:flavonoid 3-O-Glucosyltransferase, a Homologue of the Enzyme Encoded by the Maize Bronze-1 Locus That May Primarily Serve to Glucosylate Anthocyanidins in Vivo. J. Biol. Chem. 1998, 273, 9224–9233. [Google Scholar] [CrossRef] [Green Version]
- Goto-Yamamoto, N.; Wan, G.H.; Masaki, K.; Kobayashi, S. Structure and Transcription of Three Chalcone Synthase Genes of Grapevine (Vitis Vinifera). Plant Sci. 2002, 162, 867–872. [Google Scholar] [CrossRef]
- Bogs, J.; Downey, M.O.; Harvey, J.S.; Ashton, A.R.; Tanner, G.J.; Robinson, S.P. Proanthocyanidin Synthesis and Expression of Genes Encoding Leucoanthocyanidin Reductase and Anthocyanidin Reductase in Developing Grape Berries and Grapevine Leaves. Plant Physiol. 2005, 139, 652–663. [Google Scholar] [CrossRef] [Green Version]
- Bogs, J.; Ebadi, A.; McDavid, D.; Robinson, S.P. Identification of the Flavonoid Hydroxylases from Grapevine and Their Regulation during Fruit Development. Plant Physiol. 2006, 140, 279–291. [Google Scholar] [CrossRef] [Green Version]
- Fujita, A.; Goto-Yamamoto, N.; Aramaki, I.; Hashizume, K. Organ-Specific Transcription of Putative Flavonol Synthase Genes of Grapevine and Effects of Plant Hormones and Shading on Flavonol Biosynthesis in Grape Berry Skins. Biosci. Biotechnol. Biochem. 2006, 70, 632–638. [Google Scholar] [CrossRef] [PubMed]
- Petit, P.; Granier, T.; d’Estaintot, B.L.; Manigand, C.; Bathany, K.; Schmitter, J.-M.; Lauvergeat, V.; Hamdi, S.; Gallois, B. Crystal Structure of Grape Dihydroflavonol 4-Reductase, a Key Enzyme in Flavonoid Biosynthesis. J. Mol. Biol. 2007, 368, 1345–1357. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.; Curtin, C.; Bézier, A.; Franco, C.; Zhang, W. Purification, Molecular Cloning, and Characterization of Glutathione S-Transferases (GSTs) from Pigmented Vitis Vinifera L. Cell Suspension Cultures as Putative Anthocyanin Transport Proteins. J. Exp. Bot. 2008, 59, 3621–3634. [Google Scholar] [CrossRef] [Green Version]
- Gomez, C.; Terrier, N.; Torregrosa, L.; Vialet, S.; Fournier-Level, A.; Verriès, C.; Souquet, J.-M.; Mazauric, J.-P.; Klein, M.; Cheynier, V.; et al. Grapevine MATE-Type Proteins Act as Vacuolar H+-Dependent Acylated Anthocyanin Transporters. Plant Physiol. 2009, 150, 402–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hugueney, P.; Provenzano, S.; Verriès, C.; Ferrandino, A.; Meudec, E.; Batelli, G.; Merdinoglu, D.; Cheynier, V.; Schubert, A.; Ageorges, A. A Novel Cation-Dependent O-Methyltransferase Involved in Anthocyanin Methylation in Grapevine. Plant Physiol. 2009, 150, 2057–2070. [Google Scholar] [CrossRef] [PubMed]
- Falginella, L.; Castellarin, S.D.; Testolin, R.; Gambetta, G.A.; Morgante, M.; Di Gaspero, G. Expansion and Subfunctionalisation of Flavonoid 3’,5’-Hydroxylases in the Grapevine Lineage. BMC Genomics 2010, 11, 562. [Google Scholar] [CrossRef] [Green Version]
- Lücker, J.; Martens, S.; Lund, S.T. Characterization of a Vitis Vinifera Cv. Cabernet Sauvignon 3’,5’-O-Methyltransferase Showing Strong Preference for Anthocyanins and Glycosylated Flavonols. Phytochemistry 2010, 71, 1474–1484. [Google Scholar] [CrossRef]
- Fournier-Level, A.; Hugueney, P.; Verriès, C.; This, P.; Ageorges, A. Genetic Mechanisms Underlying the Methylation Level of Anthocyanins in Grape (Vitis vinifera L.). BMC Plant Biol. 2011, 11, 179. [Google Scholar] [CrossRef] [Green Version]
- Gomez, C.; Conejero, G.; Torregrosa, L.; Cheynier, V.; Terrier, N.; Ageorges, A. In Vivo Grapevine Anthocyanin Transport Involves Vesicle-Mediated Trafficking and the Contribution of anthoMATE Transporters and GST. Plant J. 2011, 67, 960–970. [Google Scholar] [CrossRef]
- Francisco, R.M.; Regalado, A.; Ageorges, A.; Burla, B.J.; Bassin, B.; Eisenach, C.; Zarrouk, O.; Vialet, S.; Marlin, T.; Chaves, M.M.; et al. ABCC1, an ATP Binding Cassette Protein from Grape Berry, Transports Anthocyanidin 3-O-Glucosides. Plant Cell 2013, 25, 1840–1854. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Díaz, R.; Ryngajllo, M.; Pérez-Díaz, J.; Peña-Cortés, H.; Casaretto, J.A.; González-Villanueva, E.; Ruiz-Lara, S. VvMATE1 and VvMATE2 Encode Putative Proanthocyanidin Transporters Expressed during Berry Development in Vitis Vinifera L. Plant Cell Rep. 2014, 33, 1147–1159. [Google Scholar] [CrossRef] [PubMed]
- Rinaldo, A.; Cavallini, E.; Jia, Y.; Moss, S.M.A.; McDavid, D.A.J.; Hooper, L.C.; Robinson, S.P.; Tornielli, G.B.; Zenoni, S.; Ford, C.M.; et al. A Grapevine Anthocyanin Acyltransferase, Transcriptionally Regulated by VvMYBA, Can Produce Most Acylated Anthocyanins Present in Grape Skins. Plant Physiol. 2015, 163, 1897–1916. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Díaz, R.; Madrid-Espinoza, J.; Salinas-Cornejo, J.; González-Villanueva, E.; Ruiz-Lara, S. Differential Roles for VviGST1, VviGST3, and VviGST4 in Proanthocyanidin and Anthocyanin Transport in Vitis Vinífera. Front. Plant Sci. 2016, 7, 1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boss, P.K.; Davies, C.; Robinson, S.P. Expression of Anthocyanin Biosynthesis Pathway Genes in Red and White Grapes. Plant Mol. Biol. 1996, 32, 565–569. [Google Scholar] [CrossRef]
- Bogs, J.; Jaffé, F.W.; Takos, A.M.; Walker, A.R.; Robinson, S.P. The Grapevine Transcription Factor VvMYBPA1 Regulates Proanthocyanidin Synthesis during Fruit Development. Plant Physiol. 2007, 143, 1347–1361. [Google Scholar] [CrossRef] [Green Version]
- Walker, A.R.; Lee, E.; Bogs, J.; McDavid, D.A.J.; Thomas, M.R.; Robinson, S.P. White Grapes Arose through the Mutation of Two Similar and Adjacent Regulatory Genes. Plant J. 2007, 49, 772–785. [Google Scholar] [CrossRef]
- Czemmel, S.; Stracke, R.; Weisshaar, B.; Cordon, N.; Harris, N.N.; Walker, A.R.; Robinson, S.P.; Bogs, J. The Grapevine R2R3-MYB Transcription Factor VvMYBF1 Regulates Flavonol Synthesis in Developing Grape Berries. Plant Physiol. 2009, 151, 1513–1530. [Google Scholar] [CrossRef] [Green Version]
- Hichri, I.; Heppel, S.C.; Pillet, J.; Léon, C.; Czemmel, S.; Delrot, S.; Lauvergeat, V.; Bogs, J. The Basic Helix-Loop-Helix Transcription Factor MYC1 Is Involved in the Regulation of the Flavonoid Biosynthesis Pathway in Grapevine. Mol. Plant 2010, 3, 509–523. [Google Scholar] [CrossRef]
- Cavallini, E.; Zenoni, S.; Finezzo, L.; Guzzo, F.; Zamboni, A.; Avesani, L.; Tornielli, G.B. Functional Diversification of Grapevine MYB5a and MYB5b in the Control of Flavonoid Biosynthesis in a Petunia Anthocyanin Regulatory Mutant. Plant Cell Physiol. 2014, 55, 517–534. [Google Scholar] [CrossRef]
- Koyama, K.; Numata, M.; Nakajima, I.; Goto-Yamamoto, N.; Matsumura, H.; Tanaka, N. Functional Characterization of a New Grapevine MYB Transcription Factor and Regulation of Proanthocyanidin Biosynthesis in Grapes. J. Exp. Bot. 2014, 65, 4433–4449. [Google Scholar] [CrossRef] [Green Version]
- Cavallini, E.; Matus, J.T.; Finezzo, L.; Zenoni, S.; Loyola, R.; Guzzo, F.; Schlechter, R.; Ageorges, A.; Arce-Johnson, P.; Tornielli, G.B. The Phenylpropanoid Pathway Is Controlled at Different Branches by a Set of R2R3-MYB C2 Repressors in Grapevine. Plant Physiol. 2015, 167, 1448–1470. [Google Scholar] [CrossRef] [PubMed]
- Matus, J.T.; Cavallini, E.; Loyola, R.; Höll, J.; Finezzo, L.; Dal Santo, S.; Vialet, S.; Commisso, M.; Roman, F.; Schubert, A.; et al. A Group of Grapevine MYBA Transcription Factors Located in Chromosome 14 Control Anthocyanin Synthesis in Vegetative Organs with Different Specificities Compared with the Berry Color Locus. Plant J. 2017, 91, 220–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, S.; Qiao, X.; Chen, H.; Nan, H.; Zhang, Z. Coordinated Regulation of Grape Berry Flesh Color by Transcriptional Activators and Repressors. J. Agric. Food Chem. 2019, 67, 11815–11824. [Google Scholar] [CrossRef] [PubMed]
- Law, J.A.; Jacobsen, S.E. Establishing, Maintaining and Modifying DNA Methylation Patterns in Plants and Animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef]
- Lei, M.; Zhang, H.; Julian, R.; Tang, K.; Xie, S.; Zhu, J.-K. Regulatory Link between DNA Methylation and Active Demethylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, 3553–3557. [Google Scholar] [CrossRef] [Green Version]
- Williams, B.P.; Pignatta, D.; Henikoff, S.; Gehring, M. Methylation-Sensitive Expression of a DNA Demethylase Gene Serves as an Epigenetic Rheostat. PLoS Genet. 2015, 11, e1005142. [Google Scholar] [CrossRef] [Green Version]
- López Sánchez, A.; Stassen, J.H.M.; Furci, L.; Smith, L.M.; Ton, J. The Role of DNA (de)methylation in Immune Responsiveness of Arabidopsis. Plant J. 2016, 88, 361–374. [Google Scholar] [CrossRef]
- Kumar, S.; Mohapatra, T. Dynamics of DNA Methylation and Its Functions in Plant Growth and Development. Front. Plant Sci. 2021, 12, 596236. [Google Scholar] [CrossRef]
- Guarino, F.; Cicatelli, A.; Castiglione, S.; Agius, D.R.; Orhun, G.E.; Fragkostefanakis, S.; Leclercq, J.; Dobránszki, J.; Kaiserli, E.; Lieberman-Lazarovich, M.; et al. An Epigenetic Alphabet of Crop Adaptation to Climate Change. Front. Genet. 2022, 13, 818727. [Google Scholar] [CrossRef]
- He, L.; Huang, H.; Bradai, M.; Zhao, C.; You, Y.; Ma, J.; Zhao, L.; Lozano-Durán, R.; Zhu, J.-K. DNA Methylation-Free Arabidopsis Reveals Crucial Roles of DNA Methylation in Regulating Gene Expression and Development. Nat. Commun. 2022, 13, 1335. [Google Scholar] [CrossRef]
- Liu, R.; How-Kit, A.; Stammitti, L.; Teyssier, E.; Rolin, D.; Mortain-Bertrand, A.; Halle, S.; Liu, M.; Kong, J.; Wu, C.; et al. A DEMETER-like DNA Demethylase Governs Tomato Fruit Ripening. Proc. Natl. Acad. Sci. USA 2015, 112, 10804–10809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, Z.; Wang, Y.; Tang, K.; Tang, D.; Datsenka, T.; Cheng, J.; Zhang, Y.; Handa, A.K.; Zhu, J.-K. Critical Roles of DNA Demethylation in the Activation of Ripening-Induced Genes and Inhibition of Ripening-Repressed Genes in Tomato Fruit. Proc. Natl. Acad. Sci. USA 2017, 114, E4511–E4519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Telias, A.; Lin-Wang, K.; Stevenson, D.E.; Cooney, J.M.; Hellens, R.P.; Allan, A.C.; Hoover, E.E.; Bradeen, J.M. Apple Skin Patterning Is Associated with Differential Expression of MYB10. BMC Plant Biol. 2011, 11, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Sharkawy, I.; Liang, D.; Xu, K. Transcriptome Analysis of an Apple (Malus × Domestica) Yellow Fruit Somatic Mutation Identifies a Gene Network Module Highly Associated with Anthocyanin and Epigenetic Regulation. J. Exp. Bot. 2015, 66, 7359–7376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Meng, D.; Wang, A.; Li, T.; Jiang, S.; Cong, P.; Li, T. The Methylation of the PcMYB10 Promoter Is Associated with Green-Skinned Sport in Max Red Bartlett Pear. Plant Physiol. 2013, 162, 885–896. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Wang, Y.; Sun, H.; Sun, L.; Zhang, L. Transposon-Induced Methylation of the RsMYB1 Promoter Disturbs Anthocyanin Accumulation in Red-Fleshed Radish. J. Exp. Bot. 2020, 71, 2537–2550. [Google Scholar] [CrossRef]
- Ma, C.; Jing, C.; Chang, B.; Yan, J.; Liang, B.; Liu, L.; Yang, Y.; Zhao, Z. The Effect of Promoter Methylation on MdMYB1 Expression Determines the Level of Anthocyanin Accumulation in Skins of Two Non-Red Apple Cultivars. BMC Plant Biol. 2018, 18, 108. [Google Scholar] [CrossRef]
- Sicilia, A.; Scialò, E.; Puglisi, I.; Lo Piero, A.R. Anthocyanin Biosynthesis and DNA Methylation Dynamics in Sweet Orange Fruit [ L. (Osbeck)] under Cold Stress. J. Agric. Food Chem. 2020, 68, 7024–7031. [Google Scholar] [CrossRef]
- Han, M.-L.; Yin, J.; Zhao, Y.-H.; Sun, X.-W.; Meng, J.-X.; Zhou, J.; Shen, T.; Li, H.-H.; Zhang, F. How the Color Fades from Flowers: Transcriptome Sequencing and DNA Methylation Analysis. Front. Plant Sci. 2020, 11, 576054. [Google Scholar] [CrossRef]
- Liu, X.-J.; Chuang, Y.-N.; Chiou, C.-Y.; Chin, D.-C.; Shen, F.-Q.; Yeh, K.-W. Methylation Effect on Chalcone Synthase Gene Expression Determines Anthocyanin Pigmentation in Floral Tissues of Two Oncidium Orchid Cultivars. Planta 2012, 236, 401–409. [Google Scholar] [CrossRef]
- Li, W.-F.; Ning, G.-X.; Mao, J.; Guo, Z.-G.; Zhou, Q.; Chen, B.-H. Whole-Genome DNA Methylation Patterns and Complex Associations with Gene Expression Associated with Anthocyanin Biosynthesis in Apple Fruit Skin. Planta 2019, 250, 1833–1847. [Google Scholar] [CrossRef] [PubMed]
- Baubec, T.; Pecinka, A.; Rozhon, W.; Mittelsten Scheid, O. Effective, Homogeneous and Transient Interference with Cytosine Methylation in Plant Genomic DNA by Zebularine. Plant J. 2009, 57, 542–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffin, P.T.; Niederhuth, C.E.; Schmitz, R.J. A Comparative Analysis of 5-Azacytidine- and Zebularine-Induced DNA Demethylation. G3 2016, 6, 2773–2780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marquez, V.E.; Barchi, J.J., Jr.; Kelley, J.A.; Rao, K.V.R.; Agbaria, R.; Ben-Kasus, T.; Cheng, J.C.; Yoo, C.B.; Jones, P.A. Zebularine: A Unique Molecule for an Epigenetically Based Strategy in Cancer Chemotherapy. The Magic of Its Chemistry and Biology. Nucleosides Nucleotides Nucleic Acids 2005, 24, 305–318. [Google Scholar] [CrossRef]
- Champion, C.; Guianvarc’h, D.; Sénamaud-Beaufort, C.; Jurkowska, R.Z.; Jeltsch, A.; Ponger, L.; Arimondo, P.B.; Guieysse-Peugeot, A.-L. Mechanistic Insights on the Inhibition of c5 DNA Methyltransferases by Zebularine. PLoS ONE 2010, 5, e12388. [Google Scholar] [CrossRef] [Green Version]
- Bourbousse, C.; Mestiri, I.; Zabulon, G.; Bourge, M.; Formiggini, F.; Koini, M.A.; Brown, S.C.; Fransz, P.; Bowler, C.; Barneche, F. Light Signaling Controls Nuclear Architecture Reorganization during Seedling Establishment. Proc. Natl. Acad. Sci. USA 2015, 112, E2836–E2844. [Google Scholar] [CrossRef] [Green Version]
- Boonjing, P.; Masuta, Y.; Nozawa, K.; Kato, A.; Ito, H. The Effect of Zebularine on the Heat-Activated Retrotransposon ONSEN in Arabidopsis Thaliana and Vigna Angularis. Genes Genet. Syst. 2020, 95, 165–172. [Google Scholar] [CrossRef]
- Pecinka, A.; Liu, C.-H. Drugs for Plant Chromosome and Chromatin Research. Cytogenet. Genome Res. 2014, 143, 51–59. [Google Scholar] [CrossRef]
- Orta, M.L.; Calderón-Montaño, J.M.; Domínguez, I.; Pastor, N.; Burgos-Morón, E.; López-Lázaro, M.; Cortés, F.; Mateos, S.; Helleday, T. 5-Aza-2’-Deoxycytidine Causes Replication Lesions That Require Fanconi Anemia-Dependent Homologous Recombination for Repair. Nucleic Acids Res. 2013, 41, 5827–5836. [Google Scholar] [CrossRef]
- Orta, M.L.; Pastor, N.; Burgos-Morón, E.; Domínguez, I.; Calderón-Montaño, J.M.; Huertas Castaño, C.; López-Lázaro, M.; Helleday, T.; Mateos, S. Zebularine Induces Replication-Dependent Double-Strand Breaks Which Are Preferentially Repaired by Homologous Recombination. DNA Repair 2017, 57, 116–124. [Google Scholar] [CrossRef]
- Liu, C.-H.; Finke, A.; Díaz, M.; Rozhon, W.; Poppenberger, B.; Baubec, T.; Pecinka, A. Repair of DNA Damage Induced by the Cytidine Analog Zebularine Requires ATR and ATM in Arabidopsis. Plant Cell 2015, 27, 1788–1800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowicka, A.; Tokarz, B.; Zwyrtková, J.; Dvořák Tomaštíková, E.; Procházková, K.; Ercan, U.; Finke, A.; Rozhon, W.; Poppenberger, B.; Otmar, M.; et al. Comparative Analysis of Epigenetic Inhibitors Reveals Different Degrees of Interference with Transcriptional Gene Silencing and Induction of DNA Damage. Plant J. 2020, 102, 68–84. [Google Scholar] [CrossRef] [PubMed]
- Iwase, Y.; Shiraya, T.; Takeno, K. Flowering and Dwarfism Induced by DNA Demethylation in Pharbitis Nil. Physiol. Plant. 2010, 139, 118–127. [Google Scholar] [CrossRef]
- Malik, G.; Dangwal, M.; Kapoor, S.; Kapoor, M. Role of DNA Methylation in Growth and Differentiation in Physcomitrella Patens and Characterization of Cytosine DNA Methyltransferases. FEBS J. 2012, 279, 4081–4094. [Google Scholar] [CrossRef]
- Finnegan, E.J.; Ford, B.; Wallace, X.; Pettolino, F.; Griffin, P.T.; Schmitz, R.J.; Zhang, P.; Barrero, J.M.; Hayden, M.J.; Boden, S.A.; et al. Zebularine Treatment Is Associated with Deletion of FT-B1 Leading to an Increase in Spikelet Number in Bread Wheat. Plant Cell Environ. 2018, 41, 1346–1360. [Google Scholar] [CrossRef] [PubMed]
- Decendit, A.; Mérillon, J.M. Condensed Tannin and Anthocyanin Production in Vitis Vinifera Cell Suspension Cultures. Plant Cell Rep. 1996, 15, 762–765. [Google Scholar] [CrossRef] [PubMed]
- Asgatay, S.; Champion, C.; Marloie, G.; Drujon, T.; Senamaud-Beaufort, C.; Ceccaldi, A.; Erdmann, A.; Rajavelu, A.; Schambel, P.; Jeltsch, A.; et al. Synthesis and Evaluation of Analogues of N-Phthaloyl-L-Tryptophan (RG108) as Inhibitors of DNA Methyltransferase 1. J. Med. Chem. 2014, 57, 421–434. [Google Scholar] [CrossRef]
- Soubeyrand, E.; Basteau, C.; Hilbert, G.; van Leeuwen, C.; Delrot, S.; Gomès, E. Nitrogen Supply Affects Anthocyanin Biosynthetic and Regulatory Genes in Grapevine Cv. Cabernet-Sauvignon Berries. Phytochemistry 2014, 103, 38–49. [Google Scholar] [CrossRef]
- Arrizabalaga-Arriazu, M.; Gomès, E.; Morales, F.; Irigoyen, J.J.; Pascual, I.; Hilbert, G. Impact of 2100-Projected Air Temperature, Carbon Dioxide, and Water Scarcity on Grape Primary and Secondary Metabolites of Different Cv. Tempranillo Clones. J. Agric. Food Chem. 2021, 69, 6172–6185. [Google Scholar] [CrossRef]
- Waffo Teguo, P.; Fauconneau, B.; Deffieux, G.; Huguet, F.; Vercauteren, J.; Merillon, J.M. Isolation, Identification, and Antioxidant Activity of Three Stilbene Glucosides Newly Extracted from Vitis Vinifera Cell Cultures. J. Nat. Prod. 1998, 61, 655–657. [Google Scholar] [CrossRef]
- Reid, K.E.; Olsson, N.; Schlosser, J.; Peng, F.; Lund, S.T. An Optimized Grapevine RNA Isolation Procedure and Statistical Determination of Reference Genes for Real-Time RT-PCR during Berry Development. BMC Plant Biol. 2006, 6, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, R.; Shevchenko, O.; Ma, C.; Maury, S.; Freitag, M.; Strauss, S.H. Poplars with a PtDDM1-RNAi Transgene Have Reduced DNA Methylation and Show Aberrant Post-Dormancy Morphology. Planta 2013, 237, 1483–1493. [Google Scholar] [CrossRef] [PubMed]
- Genitoni, J.; Vassaux, D.; Delaunay, A.; Citerne, S.; Portillo Lemus, L.; Etienne, M.-P.; Renault, D.; Stoeckel, S.; Barloy, D.; Maury, S. Hypomethylation of the Aquatic Invasive Plant, Ludwigia Grandiflora Subsp. Hexapetala Mimics the Adaptive Transition into the Terrestrial Morphotype. Physiol. Plant. 2020, 170, 280–298. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Tang, K.; Wang, B.; Duan, C.-G.; Lang, Z.; Zhu, J.-K. Protocol: A Beginner’s Guide to the Analysis of RNA-Directed DNA Methylation in Plants. Plant Methods 2014, 10, 18. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Bonnot, T.; Gillard, M.; Nagel, D. A Simple Protocol for Informative Visualization of Enriched Gene Ontology Terms. Bio-Protocol 2019, 9, e3429. [Google Scholar] [CrossRef]
- Lohse, M.; Nagel, A.; Herter, T.; May, P.; Schroda, M.; Zrenner, R.; Tohge, T.; Fernie, A.R.; Stitt, M.; Usadel, B. Mercator: A Fast and Simple Web Server for Genome Scale Functional Annotation of Plant Sequence Data. Plant Cell Environ. 2014, 37, 1250–1258. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Curtin, C.; Kikuchi, M.; Franco, C. Integration of Jasmonic Acid and Light Irradiation for Enhancement of Anthocyanin Biosynthesis in Vitis Vinifera Suspension Cultures. Plant Sci. 2002, 162, 459–468. [Google Scholar] [CrossRef]
- Deroles, S. Anthocyanin Biosynthesis in Plant Cell Cultures: A Potential Source of Natural Colourants. Anthocyanins 2008, 108–167. [Google Scholar] [CrossRef]
- Dédaldéchamp, F.; Uhel, C. Induction of Anthocyanin Synthesis in Nonpigmented Grape Cell Suspensions by Acting on DFR Substrate Availability or Precursors Level. Enzym. Microb. Technol. 1999, 25, 316–321. [Google Scholar] [CrossRef]
- Qu, J.-G.; Zhang, W.; Jin, M.-F.; Yu, X.-J. Effect of homogeneity on cell growth and anthocyanin biosynthesis in suspension cultures of Vitis vinifera. Sheng Wu Gong Cheng Xue Bao 2006, 22, 805–810. [Google Scholar] [CrossRef]
- Do, C.B.; Cormier, F. Effects of Low Nitrate and High Sugar Concentrations on Anthocyanin Content and Composition of Grape (Vitis Vinifera L.) Cell Suspension. Plant Cell Rep. 1991, 9, 500–504. [Google Scholar]
- Canaguier, A.; Grimplet, J.; Di Gaspero, G.; Scalabrin, S.; Duchêne, E.; Choisne, N.; Mohellibi, N.; Guichard, C.; Rombauts, S.; Le Clainche, I.; et al. A New Version of the Grapevine Reference Genome Assembly (12X.v2) and of Its Annotation (VCost.v3). Genomics Data 2017, 14, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A User-Driven Tool to Display Genomics Data Sets onto Diagrams of Metabolic Pathways and Other Biological Processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Baena-González, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A Central Integrator of Transcription Networks in Plant Stress and Energy Signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef]
- Alford, S.R.; Rangarajan, P.; Williams, P.; Gillaspy, G.E. Myo-Inositol Oxygenase Is Required for Responses to Low Energy Conditions in Arabidopsis Thaliana. Front. Plant Sci. 2012, 3, 69. [Google Scholar] [CrossRef] [Green Version]
- Höll, J.; Vannozzi, A.; Czemmel, S.; D’Onofrio, C.; Walker, A.R.; Rausch, T.; Lucchin, M.; Boss, P.K.; Dry, I.B.; Bogs, J. The R2R3-MYB Transcription Factors MYB14 and MYB15 Regulate Stilbene Biosynthesis in Vitis Vinifera. Plant Cell 2013, 25, 4135–4149. [Google Scholar] [CrossRef] [Green Version]
- Gao, S.; Ma, W.; Lyu, X.; Cao, X.; Yao, Y. Melatonin May Increase Disease Resistance and Flavonoid Biosynthesis through Effects on DNA Methylation and Gene Expression in Grape Berries. BMC Plant Biol. 2020, 20, 231. [Google Scholar] [CrossRef]
- Pereira, H.S.; Sofia Pereira, H.; Barão, A.; Delgado, M.; Morais-Cecílio, L.; Viegas, W. Genomic Analysis of Grapevine Retrotransposon 1 (Gret1) in Vitis Vinifera. Theor. Appl. Genet. 2005, 111, 871–878. [Google Scholar] [CrossRef]
- Benjak, A.; Forneck, A.; Casacuberta, J.M. Genome-Wide Analysis of the “Cut-and-Paste” Transposons of Grapevine. PLoS ONE 2008, 3, e3107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Kasus, T.; Ben-Zvi, Z.; Marquez, V.E.; Kelley, J.A.; Agbaria, R. Metabolic Activation of Zebularine, a Novel DNA Methylation Inhibitor, in Human Bladder Carcinoma Cells. Biochem. Pharmacol. 2005, 70, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Yi, D.; Kamei, C.L.A.; Cools, T.; Vanderauwera, S.; Takahashi, N.; Okushima, Y.; Eekhout, T.; Yoshiyama, K.O.; Larkin, J.; Van den Daele, H.; et al. The Arabidopsis SIAMESE-RELATED Cyclin-Dependent Kinase Inhibitors SMR5 and SMR7 Regulate the DNA Damage Checkpoint in Response to Reactive Oxygen Species. Plant Cell 2014, 26, 296–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bundock, P.; van Attikum, H.; Hooykaas, P. Increased Telomere Length and Hypersensitivity to DNA Damaging Agents in an Arabidopsis KU70 Mutant. Nucleic Acids Res. 2002, 30, 3395–3400. [Google Scholar] [CrossRef] [Green Version]
- Bouyer, D.; Heese, M.; Chen, P.; Harashima, H.; Roudier, F.; Grüttner, C.; Schnittger, A. Genome-Wide Identification of RETINOBLASTOMA RELATED 1 Binding Sites in Arabidopsis Reveals Novel DNA Damage Regulators. PLoS Genet. 2018, 14, e1007797. [Google Scholar] [CrossRef]
- Do, C.B.; Cormier, F. Accumulation of Anthocyanins Enhanced by a High Osmotic Potential in Grape (Vitis Vinifera L.) Cell Suspensions. Plant Cell Rep. 1990, 9, 143–146. [Google Scholar] [CrossRef]
- Cormier, F.; Do, C.B.; Nicolas, Y. Anthocyanin Production in Selected Cell Lines of Grape (Vitis Vinifera L.). Cell. Dev. Biol. Plant 1994, 30, 171–173. [Google Scholar] [CrossRef]
- Qu, J.; Zhang, W.; Yu, X.; Jin, M. Instability of Anthocyanin Accumulation in Vitis Vinifera L. Var. Gamay Fréaux Suspension Cultures. Biotechnol. Bioprocess Eng. 2005, 10, 155–161. [Google Scholar] [CrossRef]
- Conn, S.; Franco, C.; Zhang, W. Characterization of Anthocyanic Vacuolar Inclusions in Vitis Vinifera L. Cell Suspension Cultures. Planta 2010, 231, 1343–1360. [Google Scholar] [CrossRef]
- Saigne-Soulard, C.; Richard, T.; Mérillon, J.-M.; Monti, J.-P. 13C NMR Analysis of Polyphenol Biosynthesis in Grape Cells: Impact of Various Inducing Factors. Anal. Chim. Acta 2006, 563, 137–144. [Google Scholar] [CrossRef]
- Soubeyrand, E.; Colombié, S.; Beauvoit, B.; Dai, Z.; Cluzet, S.; Hilbert, G.; Renaud, C.; Maneta-Peyret, L.; Dieuaide-Noubhani, M.; Mérillon, J.-M.; et al. Constraint-Based Modeling Highlights Cell Energy, Redox Status and α-Ketoglutarate Availability as Metabolic Drivers for Anthocyanin Accumulation in Grape Cells Under Nitrogen Limitation. Front. Plant Sci. 2018, 9, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belhadj, A.; Telef, N.; Saigne, C.; Cluzet, S.; Barrieu, F.; Hamdi, S.; Mérillon, J.-M. Effect of Methyl Jasmonate in Combination with Carbohydrates on Gene Expression of PR Proteins, Stilbene and Anthocyanin Accumulation in Grapevine Cell Cultures. Plant Physiol. Biochem. 2008, 46, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Zhang, W.; Yu, X. A Combination of Elicitation and Precursor Feeding Leads to Increased Anthocyanin Synthesis in Cell Suspension Cultures of Vitis Vinifera. Plant Cell Tissue Organ Cult. 2011, 107, 261–269. [Google Scholar] [CrossRef]
- Martins, V.; Garcia, A.; Costa, C.; Sottomayor, M.; Gerós, H. Calcium- and Hormone-Driven Regulation of Secondary Metabolism and Cell Wall Enzymes in Grape Berry Cells. J. Plant Physiol. 2018, 231, 57–67. [Google Scholar] [CrossRef] [Green Version]
- Deluc, L.; Bogs, J.; Walker, A.R.; Ferrier, T.; Decendit, A.; Merillon, J.-M.; Robinson, S.P.; Barrieu, F. The Transcription Factor VvMYB5b Contributes to the Regulation of Anthocyanin and Proanthocyanidin Biosynthesis in Developing Grape Berries. Plant Physiol. 2008, 147, 2041–2053. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.-S.; Lin, C.-C.; Li, Y.-C.; Wu, M.-T.; Tsai, M.-H.; Hsing, Y.-I.C.; Jeng, S.-T. Interaction of Small RNA-8105 and the Intron of IbMYB1 RNA Regulates IbMYB1 Family Genes through Secondary siRNAs and DNA Methylation after Wounding. Plant J. 2013, 75, 781–794. [Google Scholar] [CrossRef]
- Sanchez-Muñoz, R.; Moyano, E.; Khojasteh, A.; Bonfill, M.; Cusido, R.M.; Palazon, J. Genomic Methylation in Plant Cell Cultures: A Barrier to the Development of Commercial Long-Term Biofactories. Eng. Life Sci. 2019, 19, 872–879. [Google Scholar] [CrossRef] [Green Version]
- Shirai, K.; Sato, M.P.; Nishi, R.; Seki, M.; Suzuki, Y.; Hanada, K. Positive Selective Sweeps of Epigenetic Mutations Regulating Specialized Metabolites in Plants. Genome Res. 2021, 31, 1060–1068. [Google Scholar] [CrossRef]
- Kooke, R.; Morgado, L.; Becker, F.; van Eekelen, H.; Hazarika, R.; Zheng, Q.; de Vos, R.C.H.; Johannes, F.; Keurentjes, J.J.B. Epigenetic Mapping of the Metabolome Reveals Mediators of the Epigenotype-Phenotype Map. Genome Res. 2019, 29, 96–106. [Google Scholar] [CrossRef] [Green Version]
- Majerová, E.; Fojtová, M.; Mozgová, I.; Bittová, M.; Fajkus, J. Hypomethylating Drugs Efficiently Decrease Cytosine Methylation in Telomeric DNA and Activate Telomerase without Affecting Telomere Lengths in Tobacco Cells. Plant Mol. Biol. 2011, 77, 371–380. [Google Scholar] [CrossRef]
- Ramel, F.; Sulmon, C.; Cabello-Hurtado, F.; Taconnat, L.; Martin-Magniette, M.-L.; Renou, J.-P.; El Amrani, A.; Couée, I.; Gouesbet, G. Genome-Wide Interacting Effects of Sucrose and Herbicide-Mediated Stress in Arabidopsis Thaliana: Novel Insights into Atrazine Toxicity and Sucrose-Induced Tolerance. BMC Genomics 2007, 8, 450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Culligan, K.M.; Robertson, C.E.; Foreman, J.; Doerner, P.; Britt, A.B. ATR and ATM Play Both Distinct and Additive Roles in Response to Ionizing Radiation. Plant J. 2006, 48, 947–961. [Google Scholar] [CrossRef] [PubMed]
- Ricaud, L.; Proux, C.; Renou, J.-P.; Pichon, O.; Fochesato, S.; Ortet, P.; Montané, M.-H. ATM-Mediated Transcriptional and Developmental Responses to Gamma-Rays in Arabidopsis. PLoS ONE 2007, 2, e430. [Google Scholar] [CrossRef] [PubMed]
- Yoshiyama, K.; Conklin, P.A.; Huefner, N.D.; Britt, A.B. Suppressor of Gamma Response 1 (SOG1) Encodes a Putative Transcription Factor Governing Multiple Responses to DNA Damage. Proc. Natl. Acad. Sci. USA 2009, 106, 12843–12848. [Google Scholar] [CrossRef] [Green Version]
- Bourbousse, C.; Vegesna, N.; Law, J.A. SOG1 Activator and MYB3R Repressors Regulate a Complex DNA Damage Network in. Proc. Natl. Acad. Sci. USA 2018, 115, E12453–E12462. [Google Scholar] [CrossRef] [Green Version]
- Ayenew, B.; Degu, A.; Manela, N.; Perl, A.; Shamir, M.O.; Fait, A. Metabolite Profiling and Transcript Analysis Reveal Specificities in the Response of a Berry Derived Cell Culture to Abiotic Stresses. Front. Plant Sci. 2015, 6, 728. [Google Scholar] [CrossRef] [Green Version]
- Castellarin, S.D.; Pfeiffer, A.; Sivilotti, P.; Degan, M.; Peterlunger, E.; DI Gaspero, G. Transcriptional Regulation of Anthocyanin Biosynthesis in Ripening Fruits of Grapevine under Seasonal Water Deficit. Plant Cell Environ. 2007, 30, 1381–1399. [Google Scholar] [CrossRef] [Green Version]
- Dixit, A.K.; Bhatnagar, D.; Kumar, V.; Rani, A.; Manjaya, J.G.; Bhatnagar, D. Gamma Irradiation Induced Enhancement in Isoflavones, Total Phenol, Anthocyanin and Antioxidant Properties of Varying Seed Coat Colored Soybean. J. Agric. Food Chem. 2010, 58, 4298–4302. [Google Scholar] [CrossRef]
- Nagata, T.; Yamada, H.; Du, Z.; Todoriki, S.; Kikuchi, S. Microarray Analysis of Genes That Respond to Gamma-Irradiation in Arabidopsis. J. Agric. Food Chem. 2005, 53, 1022–1030. [Google Scholar] [CrossRef]
- Gicquel, M.; Taconnat, L.; Renou, J.-P.; Esnault, M.-A.; Cabello-Hurtado, F. Kinetic Transcriptomic Approach Revealed Metabolic Pathways and Genotoxic-Related Changes Implied in the Arabidopsis Response to Ionising Radiations. Plant Sci. 2012, 195, 106–119. [Google Scholar] [CrossRef]
- Genix, P.; Bligny, R.; Martin, J.-B.; Douce, R. Transient Accumulation of Asparagine in Sycamore Cells after a Long Period of Sucrose Starvation. Plant Physiol. 1990, 94, 717–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brouquisse, R.; James, F.; Pradet, A.; Raymond, P. Asparagine Metabolism and Nitrogen Distribution during Protein Degradation in Sugar-Starved Maize Root Tips. Planta 1992, 188, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Brouquisse, R.; Gaudillere, J.P.; Raymond, P. Induction of a Carbon-Starvation-Related Proteolysis in Whole Maize Plants Submitted to Light/Dark Cycles and to Extended Darkness. Plant Physiol. 1998, 117, 1281–1291. [Google Scholar] [CrossRef] [Green Version]
- Ishizaki, K.; Larson, T.R.; Schauer, N.; Fernie, A.R.; Graham, I.A.; Leaver, C.J. The Critical Role of Arabidopsis Electron-Transfer Flavoprotein:ubiquinone Oxidoreductase during Dark-Induced Starvation. Plant Cell 2005, 17, 2587–2600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hildebrandt, T.M. Synthesis versus Degradation: Directions of Amino Acid Metabolism during Arabidopsis Abiotic Stress Response. Plant Mol. Biol. 2018, 98, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Tira-Umphon, A.; Roustan, J.-P.; Chervin, C. The Stimulation by Ethylene of the UDP Glucose-Flavonoid 3-O-Glucosyltransferase (UFGT) in Grape Tissues Is Independent from the MybA Transcription Factors. Vitis 2007, 46, 210–211. [Google Scholar]
- Kobayashi, S.; Ishimaru, M.; Hiraoka, K.; Honda, C. Myb-Related Genes of the Kyoho Grape (Vitis Labruscana) Regulate Anthocyanin Biosynthesis. Planta 2002, 215, 924–933. [Google Scholar] [CrossRef] [PubMed]









| Gene Identifier | Dark | Light | Function | Homologous Gene(s) | ||
|---|---|---|---|---|---|---|
| log2FC | p adj | log2FC | p adj | |||
| Vitvi12g02472 | 4.2 | 3 × 10−79 | 3.6 | 8 × 10−39 | GEX1 (unknown function) | at5g55490 |
| Vitvi13g01990 | 1.8 | 2 × 10−3 | 3.6 | 8 × 10−18 | Cyclin-dependent protein kinase inhibitor | at5g02220 (SMR4) |
| Vitvi04g01692 | 2.6 | 3 × 10−49 | 3.3 | 1 × 10−74 | DNA repair | at1g19025 |
| Vitvi17g01550 | 3.3 | 2 × 10−17 | 4.3 | 6 × 10−12 | Brassinosteroid-signaling kinase | at5g59010 (ATBSK5) |
| Vitvi19g02101 | 2.9 | 1 × 10−9 | 4.2 | 4 × 10−8 | Ferredoxin-fold anticodon-binding domain protein | at1g55790 |
| Vitvi17g00593 | 2.3 | 1 × 10−2 | 4.1 | 2 × 10−3 | Glutathione S transferase | at1g74590 (GSTU10) |
| Vitvi17g01381 | 3.8 | 2 × 10−27 | 3.2 | 1 × 10−19 | Glutathione S transferase | at2g29420 |
| Vitvi08g01112 | 3.2 | 4 × 10−5 | 2.7 | 4 × 10−5 | ABC transporters and multidrug resistance system | at2g37360; at3g53510 |
| Vitvi12g00272 | 2.8 | 5 × 10−34 | 2.9 | 1 × 10−27 | Tyrosine transaminase | at5g36160 |
| Vitvi01g01572 | 2.8 | 2 × 10−2 | 2.4 | 3 × 10−2 | AAA-ATPase | at3g50940; at2g18193 * |
| Vitvi14g00163 | 2.5 | 3 × 10−8 | 2.6 | 6 × 10−9 | Heavy metal-associated isoprenylated protein | at5g27690 |
| Vitvi08g00076 | 2.4 | 5 × 10−16 | 1.8 | 2 × 10−6 | Detoxification efflux carrier | at1g33110 * |
| Vitvi14g00332 | 2.4 | 1 × 10−2 | 2.4 | 4 × 10−5 | Geranylgeranyl diphosphate reductase | Q9ZS34 (N. tabacum) |
| Vitvi05g02234 | 1.5 | 3 × 10−2 | 2.2 | 5 × 10−4 | Disease resistance RPP8-like protein | at5g35450 (RPP8L3) |
| Vitvi03g01650 | 1.3 | 5 × 10−3 | 2.1 | 4 × 10−20 | Pathogenesis-related protein | at2g14580 |
| Vitvi03g01542 | 1.9 | 3 × 10−8 | 2.1 | 3 × 10−19 | 2-oxoglutarate and Fe-dependent oxygenase | at3g19000 |
| Vitvi09g00559 | 1.3 | 5 × 10−3 | 2.1 | 3 × 10−10 | Glyoxalase I family protein | at1g80160 * |
| Vitvi02g01446 | 1.5 | 2 × 10−12 | 2.1 | 1 × 10−7 | Heat shock protein | at4g25200 (HSP23.6) |
| Vitvi14g01439 | 2.0 | 2 × 10−16 | 1.7 | 1 × 10−12 | Retinoblastoma related protein | at3g12280 (RBR1) |
| Vitvi09g00768 | 4.0 | 2 × 10−24 | 6.9 | 1 × 10−19 | Ubiquitin E3 SCF FBOX | at5g07610 |
| Vitvi12g00255 | 2.0 | 2 × 10−5 | 3.2 | 5 × 10−9 | NAC transcription factor | at4g28500 |
| Vitvi12g01880 | 3.0 | 2 × 10−63 | 3.0 | 2 × 10−70 | Cupin (storage protein) | at1g07750 |
| Vitvi10g02406 | 2.2 | 3 × 10−2 | 3.0 | 3 × 10−2 | MYB domain transcription factor | at2g02060 |
| Vitvi05g00582 | 2.2 | 1 × 10−38 | 1.8 | 9 × 10−17 | Calcium transporting ATPase | at3g22910 |
| Vitvi18g01607 | 1.9 | 5 × 10−5 | 2.1 | 6 × 10−6 | Protein kinase | at1g54610; at5g50860 |
| Vitvi06g00621 | 1.6 | 8 × 10−4 | 2.0 | 3 × 10−13 | UDP-glycosyltransferase | at1g07250 and homologous genes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, J.; Garcia, V.; Zehraoui, E.; Stammitti, L.; Hilbert, G.; Renaud, C.; Maury, S.; Delaunay, A.; Cluzet, S.; Lecourieux, F.; et al. Zebularine, a DNA Methylation Inhibitor, Activates Anthocyanin Accumulation in Grapevine Cells. Genes 2022, 13, 1256. https://doi.org/10.3390/genes13071256
Kong J, Garcia V, Zehraoui E, Stammitti L, Hilbert G, Renaud C, Maury S, Delaunay A, Cluzet S, Lecourieux F, et al. Zebularine, a DNA Methylation Inhibitor, Activates Anthocyanin Accumulation in Grapevine Cells. Genes. 2022; 13(7):1256. https://doi.org/10.3390/genes13071256
Chicago/Turabian StyleKong, Junhua, Virginie Garcia, Enric Zehraoui, Linda Stammitti, Ghislaine Hilbert, Christel Renaud, Stéphane Maury, Alain Delaunay, Stéphanie Cluzet, Fatma Lecourieux, and et al. 2022. "Zebularine, a DNA Methylation Inhibitor, Activates Anthocyanin Accumulation in Grapevine Cells" Genes 13, no. 7: 1256. https://doi.org/10.3390/genes13071256