Genetic Analyses of Discrete Geographic Samples of a Golden Chanterelle in Canada Reveal Evidence for Recent Regional Differentiation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Genotyping
2.3. Allelic and Genotypic Diversities
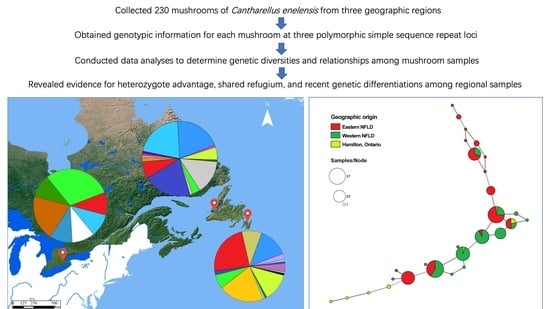
2.4. Relationships among Local and Regional Populations
2.5. STRUCTURE Analysis
2.6. Allelic Associations and Mode of Reproduction in Nature
2.7. Analyses Based on Clone-Corrected Samples
3. Results
3.1. Genetic Diversity
3.2. Distributions of Multilocus Genotypes
3.3. Hardy–Weinberg Equilibrium Test
3.4. STRUCTURE Analyses
3.5. Relationship among Local and Regional Populations in NFLD
3.6. Comparison between the NFLD Population and the Hamilton, Ontario Population
3.7. Comparison between the NFLD Populations from 2009 to 2010
4. Discussion
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thorn, R.G.; Kim, J.I.; Lebeuf, R.; Voitk, A. The golden chanterelles of Newfoundland and Labrador: A new species, a new record for North America, and a lost species rediscovered. Botany 2017, 95, 547–560. [Google Scholar] [CrossRef]
- Thorn, R.G.; Banwell, A.; Pham, T.H.; Vidal, N.P.; Manful, C.F.; Nadeem, M.; Ivanov, A.G.; Szyszka Mroz, B.; Bonneville, M.B.; Hüner, N.P.A.; et al. Identification and analyses of the chemical composition of a naturally occurring albino mutant chanterelle. Sci. Rep. 2021, 11, 20590. [Google Scholar] [CrossRef]
- Xiang, Q.P.; Wei, R.; Shao, Y.Z.; Yang, Z.Y.; Wang, X.Q.; Zhang, X.C. Phylogenetic relationships, possible ancient hybridization, and biogeographic history of Abies (Pinaceae) based on data from nuclear, plastid, and mitochondrial genomes. Mol. Phylogenet. Evol. 2015, 82, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ran, J.H.; Shen, T.T.; Liu, W.J.; Wang, P.P.; Wang, X.Q. Mitochondrial introgression and complex biogeographic history of the genus Picea. Mol. Phylogenet. Evol. 2015, 93, 63–76. [Google Scholar] [CrossRef]
- Jackson, S.T.; Webb, R.S.; Anderson, K.H.; Overpeck, J.T.; Webb, T., III; Williams, J.W.; Hansen, B.C. Vegetation and environment in eastern North America during the last glacial maximum. Quat. Sci. Rev. 2000, 19, 489–508. [Google Scholar] [CrossRef]
- Marshall, S.J.; James, T.S.; Clarke, G.K.C. North American ice sheet reconstructions at the Last Glacial Maximum. Quat. Sci. Rev. 2002, 21, 175–192. [Google Scholar] [CrossRef]
- Dunham, S.M.; Kretzer, A.; Pfrender, M.E. Characterization of Pacific golden chanterelle (Cantharellus formosus) genet size using co-dominant microsatellite markers. Mol. Ecol. 2003, 12, 1607–1618. [Google Scholar] [CrossRef] [Green Version]
- Zalapa, J.E.; Cuevas, H.; Zhu, H.; Steffan, S.; Senalik, D.; Zeldin, E.; McCown, B.; Harbut, R.; Simon, P. Using next-generation sequencing approaches to isolate simple sequence repeat (SSR) loci in the plant sciences. Am. J. Bot. 2012, 99, 193–208. [Google Scholar] [CrossRef] [Green Version]
- Broders, H.G.; Mahoney, S.P.; Montevecchi, W.A.; Davidson, W.S. Population genetic structure and the effect of founder events on the genetic variability of moose, Alces alces, in Canada. Mol. Ecol. 1999, 8, 1309–1315. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.Y.; Moon, S.; Shim, D.; Hong, C.P.; Lee, Y.; Koo, C.D.; Chung, J.W.; Ryu, H. Development of 44 novel polymorphic SSR markers for determination of shiitake mushroom (Lentinula edodes) cultivars. Genes 2017, 8, 109. [Google Scholar] [CrossRef] [Green Version]
- Korfanty, G.A.; Dixon, M.; Jia, H.; Yoell, H.; Xu, J. Genetic Diversity and Dispersal of Aspergillus fumigatus in Arctic Soils. Genes 2021, 13, 19. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Bao, H.; Bau, T. Genetic diversity and population structure of endemic mushroom Leucocalocybe mongolica in Mongolian Plateau uncovered by EST-SSR markers. Biotechnol. Biotechnol. Equip. 2018, 32, 1195–1204. [Google Scholar] [CrossRef] [Green Version]
- ArcGIS. Available online: https://www.esri.com/en-us/arcgis/ (accessed on 15 April 2022).
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Goor, R.M.; Neall, L.F.; Hoffman, D.; Sherry, S.T. A Mathematical approach to the analysis of multiplex DNA profiles. Bull. Math. Biol. 2011, 73, 1909. [Google Scholar] [CrossRef] [Green Version]
- Peakall, R.; Smouse, P.E. GenAlEx 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [Green Version]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wigginton, J.E.; Cutler, D.J.; Abecasis, G.R. A note on exact tests of Hardy-Weinberg equilibrium. Am. J. Hum. Genet. 2005, 76, 887–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergemann, S.E.; Miller, S.L. Size, distribution, and persistence of genets in local populations of the late-stage ectomycorrhizal basidiomycete, Russula brevipes. New Phytol. 2002, 156, 313–320. [Google Scholar] [CrossRef] [Green Version]
- Arnaud-Haond, S.; Duarte, C.M.; Alberto, F.; Serrão, E.A. Standardizing methods to address clonality in population studies. Mol. Ecol. 2007, 16, 5115–5139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charest, R.; Brouillet, L.; Bouchard, A.; Hay, S. The vascular flora of Terra Nova National Park, Newfoundland, Canada: A biodiversity analysis from a biogeographical and life form perspective. Can. J. Bot. 2000, 78, 629–645. [Google Scholar]
- Rajora, O.P.; DeVerno, L.; Mosseler, A.; Innes, D.J. Genetic diversity and population structure of disjunct Newfoundland and central Ontario populations of eastern white pine (Pinus strobus). Can. J. Bot. 1998, 76, 500–508. [Google Scholar]
- Mahoney, S.P.; Virgl, J.A.; Mawhinney, K. Potential mechanisms of phenotypic divergence in body size between Newfoundland and mainland black bear populations. Can. J. Zool. 2001, 79, 1650–1660. [Google Scholar] [CrossRef]
- Zhai, G.; Zhou, J.; Woods, M.O.; Green, J.S.; Parfrey, P.; Rahman, P.; Green, R.C. Genetic structure of the Newfoundland and Labrador population: Founder effects modulate variability. Eur. J. Hum. Genet. 2016, 24, 1063–1070. [Google Scholar] [CrossRef] [Green Version]
- Yao, T. The response of currents in Trinity Bay, Newfoundland, to local wind forcing. Atmos. Ocean 1986, 24, 235–252. [Google Scholar] [CrossRef]
- Deser, C.; Blackmon, M.L. Surface climate variations over the North Atlantic Ocean during winter: 1900–1989. J. Clim. 1993, 6, 1743–1753. [Google Scholar] [CrossRef]
- Prinsenberg, S.J.; Peterson, I.K.; Narayanan, S.; Umoh, J.U. Interaction between atmosphere, ice cover, and ocean off Labrador and Newfoundland from 1962 to 1992. Can. J. Fish. Aquat. Sci. 1997, 54, 30–39. [Google Scholar] [CrossRef]
- Banfield, C.E.; Jacobs, J.D. Regional patterns of temperature and precipitation for Newfoundland and Labrador during the past century. Can Geogr. 1998, 42, 354–364. [Google Scholar] [CrossRef]
- Anderson, J.T.; Gregory, R.S.; Collins, W.T. Acoustic classification of marine habitats in coastal Newfoundland. ICES Mar. Sci. Symp. 2002, 59, 156–167. [Google Scholar] [CrossRef]
- Vilneff, C.; Thorn, R.G. Newfoundland golden chanterelles: Examining their identity and regional levels of damage by slugs and larvae. Omphalina 2011, 2, 13–17. [Google Scholar]
- Sass, J.E. The cytological basis for homothallism and heterothallism in the Agaricaceae. Am. J. Bot. 1929, 16, 663–701. [Google Scholar] [CrossRef]
- Foulongne-Oriol, M.; Taskent, O.; Kües, U.; Sonnenberg, A.S.; van Peer, A.F.; Giraud, T. Mating-type locus organization and mating-type chromosome differentiation in the bipolar edible button mushroom Agaricus bisporus. Genes 2021, 12, 1079. [Google Scholar] [CrossRef]
- Zhang, P.; Tang, L.P.; Cai, Q.; Xu, J. A review on the diversity, phylogeography and population genetics of Amanita mushrooms. Mycology 2015, 6, 86–93. [Google Scholar] [CrossRef] [Green Version]
- Raper, C.A.; Raper, J.R.; Miller, R.E. Genetic analysis of the life cycle of Agaricus bisporus. Mycologia 1972, 64, 1088–1117. [Google Scholar] [CrossRef]
- Xu, J.P.; Desmerger, C.; Callac, P. Fine-scale genetic analyses reveal unexpected spatial-temporal heterogeneity in two natural populations of the commercial mushroom Agaricus bisporus. Microbiology 2002, 148, 1253–1262. [Google Scholar] [CrossRef] [Green Version]
- Pizarro, D.; Dal Grande, F.; Leavitt, S.D.; Dyer, P.S.; Schmitt, I.; Crespo, A.; Lumbsch, H.T.; Divakar, P.K. Whole-genome sequence data uncover widespread heterothallism in the largest group of lichen-forming fungi. Genome Biol. Evol. 2019, 11, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Danell, E.; Fries, N. Methods for isolation of Cantharellus species, and the synthesis of ectomycorrhizae with Picea abies. Mycotaxon 1990, 38, 141–148. [Google Scholar]
- Danell, E. Formation and growth of the ectomycorrhiza of Cantharellus cibarius. Mycorrhiza 1994, 5, 89–97. [Google Scholar] [CrossRef]
- Pöggeler, S. Mating-type genes for classical strain improvements of ascomycetes. Appl. Microbiol. Biotechnol. 2001, 56, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Rahman, P.; Jones, A.; Curtis, J.; Bartlett, S.; Peddle, L.; Fernandez, B.A.; Freimer, N.B. The Newfoundland population: A unique resource for genetic investigation of complex diseases. Hum. Mol. Genet. 2003, 12 (Suppl. 2), R167–R172. [Google Scholar] [CrossRef] [Green Version]
- Ruzzante, D.E.; Taggart, C.T.; Doyle, R.W.; Cook, D. Stability in the historical pattern of genetic structure of Newfoundland cod (Gadus morhua) despite the catastrophic decline in population size from 1964 to 1994. Conserv. Genet. 2001, 2, 257–269. [Google Scholar] [CrossRef]
- Thorn, R.G.; Voitk, A. The Newfoundland chanterelle. Omphalina 2011, 2, 14–17. [Google Scholar]
- Horton, T.R.; Bruns, T.D. The molecular revolution in ectomycorrhizal ecology: Peeking into the black-box. Mol. Ecol. 2001, 10, 1855–1871. [Google Scholar] [CrossRef] [Green Version]
- Tedersoo, L.; Mett, M.; Ishida, T.A.; Bahram, M. Phylogenetic relationships among host plants explain differences in fungal species richness and community composition in ectomycorrhizal symbiosis. New Phytol. 2013, 199, 822–831. [Google Scholar] [CrossRef]


| Population | Locus | N | Na | Ne | uh |
|---|---|---|---|---|---|
| Eastern NFLD | Cf113 | 110 | 4 | 1.78 | 0.44 |
| Cf126 | 110 | 3 | 2.19 | 0.55 | |
| Cf642 | 110 | 2 | 1.54 | 0.35 | |
| Mean | 110 | 3 | 1.84 | 0.45 | |
| Standard Error | 0 | 0.58 | 0.19 | 0.06 | |
| Western NFLD | Cf113 | 110 | 3 | 1.96 | 0.49 |
| Cf126 | 110 | 3 | 1.55 | 0.36 | |
| Cf642 | 110 | 2 | 1.07 | 0.06 | |
| Mean | 110 | 2.67 | 1.53 | 0.30 | |
| Standard Error | 0 | 0.33 | 0.26 | 0.13 | |
| Hamilton, Ontario | Cf113 | 10 | 3 | 2.25 | 0.58 |
| Cf126 | 10 | 2 | 1.98 | 0.52 | |
| Cf642 | 10 | 2 | 1.73 | 0.44 | |
| Mean | 10 | 2.33 | 1.98 | 0.52 | |
| Standard Error | 0 | 0.33 | 0.15 | 0.04 |
| Region | Locus | Nonclone-Corrected | Clone-Corrected | ||||
|---|---|---|---|---|---|---|---|
| Observed Heterozygotes | Expected Heterozygotes | Chi-Square Value | Observed Heterozygotes | Expected Heterozygotes | Chi-Square Value | ||
| Eastern NFLD | Cf113 | 52 | 20.7 | 28.504 *** | 12 | 12.8 | 8.214 |
| Cf126 | 103 | 59.7 | 85.250 *** | 21 | 13.8 | 11.931 ** | |
| Cf642 | 50 | 38.6 | 9.516 ** | 14 | 10.2 | 3.529 | |
| Western NFLD | Cf113 | 71 | 51.0 | 19.395 *** | 18 | 14.8 | 2.725 |
| Cf126 | 42 | 38.9 | 0.984 | 12 | 13.4 | 0.964 | |
| Cf642 | 7 | 6.8 | 0.119 | 3 | 2.8 | 0.086 | |
| Hamilton, Ontario | Cf113 | 8 | 4.8 | 4.444 | 5 | 1.7 | 2.160 |
| Cf126 | 9 | 5.4 | 6.694 ** | 6 | 3.4 | 3.938 * | |
| Cf642 | 4 | 4.2 | 0.023 | 3 | 3.2 | 0.031 | |
| Data | Source | df | SS | MS | Est. Var. | % | Value | p |
|---|---|---|---|---|---|---|---|---|
| Nonclone-corrected | Among Regions | 1 | 351.580 | 351.580 | 2.462 | 16% | 0.160 | 0.001 *** |
| Among Local Pops | 8 | 843.160 | 105.395 | 4.865 | 32% | 0.375 | 0.001 *** | |
| Within Local Pops | 190 | 1539.000 | 8.100 | 8.100 | 53% | 0.475 | 0.001 *** | |
| Total | 199 | 2733.740 | 15.427 | 100% | ||||
| Clone-corrected | Among Regions | 1 | 98.587 | 98.587 | 3.505 | 19% | 0.195 | 0.001 *** |
| Among Local Pops | 7 | 93.576 | 13.368 | 0.000 | 0% | −0.018 | 0.583 | |
| Within Local Pops | 40 | 587.143 | 14.679 | 14.679 | 81% | 0.181 | 0.004 ** | |
| Total | 48 | 779.306 | 18.183 | 100% |
| Locus | C. formosus [7] | C. enelensis [This Study] | ||||
|---|---|---|---|---|---|---|
| Allele No. | Repeat Motif | Fragment Length * | Allele No. | Repeat Motif | Fragment Length | |
| Cf113 | 4 | (ggt)5 | 96, –, 117, 120 | 4 | (gac)4N38(acc)5 | 111, 113, 116, 119 |
| Cf126 | 3 | (ggt)6 | 221, 228, 231 | 4 | (ggt)7 | 213, 216, 219, 222 |
| Cf642 | 4 | (gaca)9 | 275, 276, 277, 279 | 3 | (gtct)4N9(tgtt)3 | 267, 290, 294 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, K.; Korfanty, G.A.; Xu, J.; Thorn, R.G. Genetic Analyses of Discrete Geographic Samples of a Golden Chanterelle in Canada Reveal Evidence for Recent Regional Differentiation. Genes 2022, 13, 1110. https://doi.org/10.3390/genes13071110
Zhao K, Korfanty GA, Xu J, Thorn RG. Genetic Analyses of Discrete Geographic Samples of a Golden Chanterelle in Canada Reveal Evidence for Recent Regional Differentiation. Genes. 2022; 13(7):1110. https://doi.org/10.3390/genes13071110
Chicago/Turabian StyleZhao, Kuan, Gregory A. Korfanty, Jianping Xu, and R. Greg Thorn. 2022. "Genetic Analyses of Discrete Geographic Samples of a Golden Chanterelle in Canada Reveal Evidence for Recent Regional Differentiation" Genes 13, no. 7: 1110. https://doi.org/10.3390/genes13071110