Characterization and Functional Study of FAM49B Reveals Its Effect on Cell Proliferation in HEK293T Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. RNA Extraction and Reverse Transcription-PCR
2.3. Western Blot and Cell Immunofluorescence
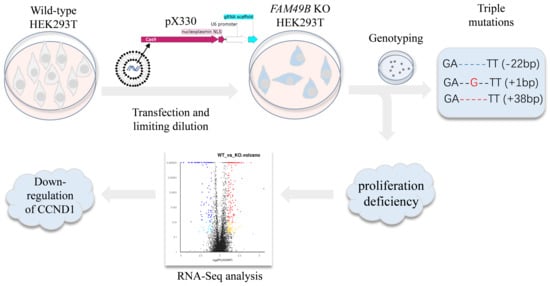
2.4. Genome Editing FAM49B in HEK293T Cells
2.5. Genotyping of Mutant Single-Cell Lines
2.6. Cell Proliferation Assay
2.7. Cell Cycle Analysis
2.8. Apoptosis Detection
2.9. RNA-Seq and Data Analysis
2.10. Statistical Analysis
3. Results
3.1. Characterization of FAM49B in HEK293T Cells
3.2. Establishing FAM49B Mutant HEK293T Cell Line
3.3. Triple Alleles of FAM49B Exist in HEK293T Genome
3.4. FAM49B KO Cells Exhibit Proliferation Deficiency
3.5. FAM49B Mutant HEK293T Cells Display No Remarkable Differences in Cell Cycle Arrests or Apoptosis
3.6. The Alternative Splicing of FAM49B in HEK293T
3.7. CCND1 Expression Is Dramatically Decreased in FAM49B Mutant Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Z.; Borek, D.; Padrick, S.B.; Gomez, T.S.; Metlagel, Z.; Ismail, A.M.; Umetani, J.; Billadeau, D.D.; Otwinowski, Z.; Rosen, M.K. Structure and control of the actin regulatory WAVE complex. Nature 2010, 468, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Le, A.H.; Yelland, T.; Paul, N.R.; Fort, L.; Nikolaou, S.; Ismail, S.; Machesky, L.M. CYRI-A limits invasive migration through macropinosome formation and integrin uptake regulation. J. Cell Biol. 2021, 220, e202012114. [Google Scholar] [CrossRef] [PubMed]
- El Masri, R.; Delon, J. RHO GTPases: From new partners to complex immune syndromes. Nat. Rev. Immunol. 2021, 21, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Kaplan, E.; Stone, R.; Hume, P.J.; Greene, N.P.; Koronakis, V. Structure of CYRI-B (FAM49B), a key regulator of cellular actin assembly. Acta Crystallogr. Sect. D Struct. Biol. 2020, 76, 1015–1024. [Google Scholar] [CrossRef]
- Yelland, T.; Le, A.H.; Nikolaou, S.; Insall, R.; Machesky, L.; Ismail, S. Structural Basis of CYRI-B Direct Competition with Scar/WAVE Complex for Rac1. Structure 2021, 29, 226–237. [Google Scholar] [CrossRef]
- Schenck, A.; Bardoni, B.; Langmann, C.; Harden, N.; Mandel, J.; Giangrande, A. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron 2003, 38, 887–898. [Google Scholar] [CrossRef] [Green Version]
- Fort, L.; Batista, J.M.; Thomason, P.A.; Spence, H.J.; Whitelaw, J.A.; Tweedy, L.; Greaves, J.; Martin, K.J.; Anderson, K.I.; Brown, P. Fam49/CYRI interacts with Rac1 and locally suppresses protrusions. Nat. Cell Biol. 2018, 20, 1159–1171. [Google Scholar] [CrossRef] [Green Version]
- Whitelaw, J.A.; Lilla, S.; Paul, N.R.; Fort, L.; Zanivan, S.; Machesky, L.M. CYRI/Fam49 proteins represent a new class of Rac1 interactors. Commun. Integr. Biol. 2019, 12, 112–118. [Google Scholar] [CrossRef]
- Yuki, K.E.; Marei, H.; Fiskin, E.; Eva, M.M.; Gopal, A.A.; Schwartzentruber, J.A.; Majewski, J.; Cellier, M.; Mandl, J.N.; Vidal, S.M.; et al. CYRI/FAM49B negatively regulates RAC1-driven cytoskeletal remodelling and protects against bacterial infection. Nat. Microbiol. 2019, 4, 1516–1531. [Google Scholar] [CrossRef]
- Li, W.; Guan, X.; Jiang, S.; Sun, L. The novel fish miRNA pol-miR-novel_171 and its target gene FAM49B play a critical role in apoptosis and bacterial infection. Dev. Comp. Immunol. 2020, 106, 103616. [Google Scholar] [CrossRef]
- Nagarajan, N.A.; Gonzalez, F.; Shastri, N. Nonclassical MHC class Ib–restricted cytotoxic T cells monitor antigen processing in the endoplasmic reticulum. Nat. Immunol. 2012, 13, 579–586. [Google Scholar] [CrossRef] [Green Version]
- Kim, J. Functional Characterization of Fam49b in T Cells during Salmonella Infection; McGill University: Montreal, QC, Canada, 2020; ISBN 9798708707581. [Google Scholar]
- Shang, W.; Jiang, Y.; Boettcher, M.; Ding, K.; Mollenauer, M.; Liu, Z.; Wen, X.; Liu, C.; Hao, P.; Zhao, S.; et al. Genome-wide CRISPR screen identifies FAM49B as a key regulator of actin dynamics and T cell activation. Proc. Natl. Acad. Sci. USA 2018, 115, E4051–E4060. [Google Scholar] [CrossRef] [Green Version]
- Gilli, F.; Lindberg, R.L.P.; Valentino, P.; Marnetto, F.; Malucchi, S.; Sala, A.; Capobianco, M.; di Sapio, A.; Sperli, F.; Kappos, L.; et al. Learning from nature: Pregnancy changes the expression of inflammation-related genes in patients with multiple sclerosis. PLoS ONE 2010, 5, e8962. [Google Scholar] [CrossRef]
- Gilli, F.; Navone, N.D.; Perga, S.; Marnetto, F.; Caldano, M.; Capobianco, M.; Pulizzi, A.; Malucchi, S.; Bertolotto, A. Loss of braking signals during inflammation: A factor affecting the development and disease course of multiple sclerosis. Arch. Neurol. 2011, 68, 879–888. [Google Scholar] [CrossRef] [Green Version]
- Chattaragada, M.S.; Riganti, C.; Sassoe, M.; Principe, M.; Santamorena, M.M.; Roux, C.; Curcio, C.; Evangelista, A.; Allavena, P.; Salvia, R.; et al. FAM49B, a novel regulator of mitochondrial function and integrity that suppresses tumor metastasis. Oncogene 2018, 37, 697–709. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Xiong, Y.; Wang, Z.; Han, J.; Shi, S.; He, J.; Shen, N.; Wu, W.; Wang, R.; Lv, W. FAM49B Promotes Breast Cancer Proliferation, Metastasis, and Chemoresistance by Stabilizing ELAVL1 Protein and Regulating Downstream Rab10/TLR4 Pathway. Cancer Cell Int. 2021, 21, 1–20. [Google Scholar] [CrossRef]
- Zhang, Y.; Du, P.; Li, Y.; Zhu, Q.; Song, X.; Liu, S.; Hao, J.; Liu, L.; Liu, F.; Hu, Y.; et al. TASP1 Promotes Gallbladder Cancer Cell Proliferation and Metastasis by Up-regulating FAM49B via PI3K/AKT Pathway. Int. J. Biol. Sci. 2020, 16, 739–751. [Google Scholar] [CrossRef] [Green Version]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, P.; Chatterjee, M. Antiproliferative role of vitamin D and its analogs–a brief overview. Mol. Cell. Biochem. 2003, 253, 247–254. [Google Scholar] [CrossRef]
- Guo, M.; Hay, B.A. Cell proliferation and apoptosis. Curr. Opin. Cell Biol. 1999, 11, 745–752. [Google Scholar] [CrossRef]
- George, S.J.; Dwivedi, A. MMPs, cadherins, and cell proliferation. Trends Cardiovas. Med. 2004, 14, 100–105. [Google Scholar] [CrossRef]
- Bastide, P.; Darido, C.; Pannequin, J.; Kist, R.; Robine, S.; Marty-Double, C.; Bibeau, F.; Scherer, G.; Joubert, D.; Hollande, F. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J. Cell Biol. 2007, 178, 635–648. [Google Scholar] [CrossRef]
- Han, Y.; Yang, Y.; Yuan, H.; Zhang, T.; Sui, H.; Wei, X.; Liu, L.; Huang, P.; Zhang, W.; Bai, Y. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology 2014, 46, 396–401. [Google Scholar] [CrossRef]
- Sherr, C.J.; Roberts, J.M. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 1999, 13, 1501–1512. [Google Scholar] [CrossRef] [Green Version]
- Harbour, J.W.; Dean, D.C. The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes Dev. 2000, 14, 2393–2409. [Google Scholar] [CrossRef] [Green Version]
- Bartek, J.; Bartkova, J.; Lukas, J. The retinoblastoma protein pathway and the restriction point. Curr. Opin. Cell Biol. 1996, 8, 805–814. [Google Scholar] [CrossRef]
- Louis, N.; Evelegh, C.; Graham, F.L. Cloning and sequencing of the cellular–viral junctions from the human adenovirus type 5 transformed 293 cell line. Virology 1997, 233, 423–429. [Google Scholar] [CrossRef] [Green Version]
- Frisch, S.M.; Mymryk, J.S. Adenovirus-5 E1A: Paradox and paradigm. Nat. Rev. Mol. Cell Biol. 2002, 3, 441–452. [Google Scholar] [CrossRef]
- Whyte, P.; Williamson, N.M.; Harlow, E.D. Cellular targets for transformation by the adenovirus E1A proteins. Cell 1989, 56, 67–75. [Google Scholar] [CrossRef]
- Nevins, J.R. E2F: A link between the Rb tumor suppressor protein and viral oncoproteins. Science 1992, 258, 424–429. [Google Scholar] [CrossRef] [PubMed]
- Heng, Y.; Koh, C. Actin cytoskeleton dynamics and the cell division cycle. Int. J. Biochem. Cell Biol. 2010, 42, 1622–1633. [Google Scholar] [CrossRef] [PubMed]
- Brunet, S.; Maro, B. Cytoskeleton and cell cycle control during meiotic maturation of the mouse oocyte: Integrating time and space. Reproduction 2005, 130, 801–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Assoian, R.K.; Zhu, X. Cell anchorage and the cytoskeleton as partners in growth factor dependent cell cycle progression. Curr. Opin. Cell Biol. 1997, 9, 93–98. [Google Scholar] [CrossRef]
- Nakaseko, Y.; Yanagida, M. Cytoskeleton in the cell cycle. Nature 2001, 412, 291–292. [Google Scholar] [CrossRef]
- Jones, M.C.; Zha, J.; Humphries, M.J. Connections between the cell cycle, cell adhesion and the cytoskeleton. Philos. Trans. R. Soc. B 2019, 374, 20180227. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Jiang, Y.; Lao, J.; Zhou, Y.; Su, L.; Huang, X. Characterization and Functional Study of FAM49B Reveals Its Effect on Cell Proliferation in HEK293T Cells. Genes 2022, 13, 388. https://doi.org/10.3390/genes13020388
Chen Y, Jiang Y, Lao J, Zhou Y, Su L, Huang X. Characterization and Functional Study of FAM49B Reveals Its Effect on Cell Proliferation in HEK293T Cells. Genes. 2022; 13(2):388. https://doi.org/10.3390/genes13020388
Chicago/Turabian StyleChen, Yijian, Yuyan Jiang, Jihui Lao, Yankuan Zhou, Lida Su, and Xiao Huang. 2022. "Characterization and Functional Study of FAM49B Reveals Its Effect on Cell Proliferation in HEK293T Cells" Genes 13, no. 2: 388. https://doi.org/10.3390/genes13020388