Bioinformatic Characterization and Molecular Evolution of the Lucina pectinata Hemoglobins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioinformatic Analyses
2.2. Phylogenetic Analysis
2.3. Metric Multidimensional Scaling Analysis
2.4. Coding Region Alignments
3. Results
3.1. Multiple Sequence Alignment
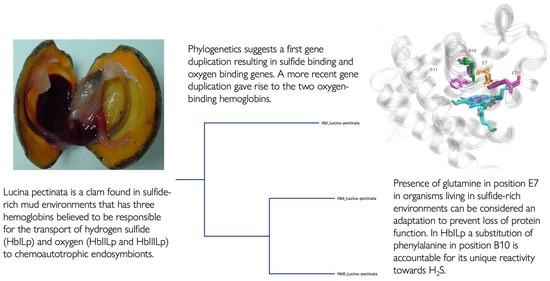
3.2. Phylogenetic Analyses
3.3. Metric Multi-Dimensional Scaling Analysis
3.4. Coding Region Bioinformatic Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weber, R.E.; Vinogradov, S.N. Nonvertebrate hemoglobins: Functions and molecular adaptations. Physiol. Rev. 2001, 81, 569–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodman, M.; Pedwaydon, J.; Czelusniak, J.; Suzuki, T.; Gotoh, T.; Moens, L.; Shishikura, F.; Walz, D.; Vinogradov, S. An evolutionary tree for invertebrate globin sequences. J. Mol. Evol. 1988, 27, 236–249. [Google Scholar] [CrossRef] [PubMed]
- Edmundson, A.B.; Hirs, C.H. The amino-acid sequence of sperm whale myoglobin. Chemical studies. Nature 1961, 190, 663–665. [Google Scholar] [CrossRef]
- Perutz, M.F. Structure and function of haemoglobin: I. A tentative atomic model of horse oxyhaemoglobin. J. Mol. Biol. 1965, 13, 646–668. [Google Scholar] [CrossRef]
- Perutz, M.F.; Muirhead, H.; Cox, J.M.; Goaman, L.C. Three-dimensional Fourier synthesis of horse oxyhaemoglobin at 2.8 A resolution: The atomic model. Nature 1968, 219, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Perutz, M.F. Regulation of oxygen affinity of hemoglobin: Influence of structure of the globin on the heme iron. Annu. Rev. Biochem. 1979, 48, 327–386. [Google Scholar] [CrossRef]
- Bashford, D.; Chothia, C.; Lesk, A.M. Determinants of a protein fold. Unique features of the globin amino acid sequences. J. Mol. Biol. 1987, 196, 199–216. [Google Scholar] [CrossRef]
- Lesk, A.M.; Chothia, C. How different amino acid sequences determine similar protein structures: The structure and evolutionary dynamics of the globins. J. Mol. Biol. 1980, 136, 225–270. [Google Scholar] [CrossRef]
- Childress, J.J. Uptake and transport of sulfide in marine invertebrates. In Comparative Physiology: Life in Water and on Land; Dejours, P., Bolis, L., Taylor, C.R., Weibel, E.R., Eds.; Springer: Berlin/Heidelberg, Germany, 1987; Volume 9, pp. 231–239. [Google Scholar]
- Felbeck, H.; Childress, J.J.; Somero, G.N. Calvin-Benson Cycle and sulphide oxidation enzymes in animals from sulphide-rich habitats. Nature 1981, 293, 291–293. [Google Scholar] [CrossRef]
- Kim, Y.W.; Yasuda, M.; Yamagishi, A.; Oshima, T.; Ohta, S. Characterization of the endosymbiont of a deep-sea bivalve, Calyptogena soyoae. Appl. Environ. Microbiol. 1995, 61, 823–827. [Google Scholar] [CrossRef]
- Chatfield, M.J.; La Mar, G.N.; Kauten, R.J. Proton NMR characterization of isomeric sulfmyoglobins: Preparation, interconversion, reactivity patterns, and structural features. Biochemistry 1987, 26, 6939–6950. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.D.; Glover, E.A. Functional anatomy, chemosymbiosis and evolution of the Lucinidae. Geol. Soc. London, Spéc. Publ. 2000, 177, 207–225. [Google Scholar] [CrossRef]
- Kraus, D.W.; Wittenberg, J.B. Hemoglobins of the Lucina pectinata/bacteria symbiosis. I. Molecular properties, kinetics and equilibria of reactions with ligands. J. Biol. Chem. 1990, 265, 16043–16053. [Google Scholar] [CrossRef]
- Rizzi, M.; Wittenberg, J.B.; Coda, A.; Fasano, M.; Ascenzi, P.; Bolognesi, M. Structure of the sulfide-reactive hemoglobin from the clam Lucina pectinata. Crystallographic analysis at 1.5 A resolution. J. Mol. Biol. 1994, 244, 86–99. [Google Scholar] [CrossRef]
- Gibson, Q.H.; Wittenberg, J.B.; Wittenberg, B.A.; Bogusz, D.; Appleby, C.A. The kinetics of ligand binding to plant hemoglobins. Structural implications. J. Biol. Chem. 1989, 264, 100–107. [Google Scholar] [CrossRef]
- Rizzi, M.; Wittenberg, J.B.; Coda, A.; Ascenzi, P.; Bolognesi, M. Structural bases for sulfide recognition in Lucina pectinata hemoglobin I. J. Mol. Biol. 1996, 258, 1–5. [Google Scholar] [CrossRef]
- Kraus, D.W.; Wittenberg, J.B.; Lu, J.F.; Peisach, J. Hemoglobins of the Lucina pectinata/bacteria symbiosis. II. An electron paramagnetic resonance and optical spectral study of the ferric proteins. J. Biol. Chem. 1990, 265, 16054–16059. [Google Scholar] [CrossRef]
- Gavira, J.A.; Camara-Artigas, A.; De Jesús-Bonilla, W.; López-Garriga, J.; Lewis, A.; Pietri, R.; García-Ruiz, J.M. Structure and ligand selection of hemoglobin II from Lucina pectinata. J. Biol. Chem. 2008, 283, 9414–9423. [Google Scholar] [CrossRef] [Green Version]
- Pietri, R.; León, R.G.; Kiger, L.; Marden, M.C.; Granell, L.B.; Cadilla, C.L.; López-Garriga, J. Hemoglobin I from Lucina pectinata: A model for distal heme-ligand control. Biochim. Biophys. Acta 2006, 1764, 758–765. [Google Scholar] [CrossRef]
- Pietri, R.; Lewis, A.; León, R.G.; Casabona, G.; Kiger, L.; Yeh, S.R.; López-Garriga, J. Factors controlling the reactivity of hydrogen sulfide with hemeproteins. Biochemistry 2009, 48, 4881–4894. [Google Scholar] [CrossRef]
- Ramos-Alvarez, C.; Yoo, B.K.; Pietri, R.; Lamarre, I.; Martin, J.L.; Lopez-Garriga, J.; Negrerie, M. Reactivity and dynamics of H2S, NO, and O2 interacting with hemoglobins from Lucina pectinata. Biochemistry 2013, 52, 7007–7021. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Martínez, C.R.; Nieves-Marrero, C.A.; Estremera-Andújar, R.A.; Gavira, J.A.; González-Ramírez, L.A.; López-Garriga, J.; García-Ruiz, J.M. Crystallization and diffraction patterns of the oxy and cyano forms of the Lucina pectinata haemoglobins complex. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008, 65 Pt 1, 25–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benson, D.A.; Karsch-Mizrachi, I.; Clark, K.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2012, 40, D48–D53. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Taly, J.F.; Magis, C.; Bussotti, G.; Chang, J.M.; Di Tommaso, P.; Erb, I.; Notredame, C. Using the T-Coffee package to build multiple sequence alignments of protein, RNA, DNA sequences and 3D structures. Nat. Protoc. 2011, 6, 1669–1682. [Google Scholar] [CrossRef]
- Nicholas, K.B.; Nicholas, H.B.; Deerfield II, D.W. GeneDoc: Analysis and Visualization of Genetic Variation. EMBNEW News 1997, 4, 14. [Google Scholar]
- Roberts, E.; Eargle, J.; Wright, D.; Luthey-Schulten, Z. MultiSeq: Unifying sequence and structure data for evolutionary analysis. BMC Bioinform. 2006, 7, 382. [Google Scholar] [CrossRef] [Green Version]
- Russell, R.B.; Barton, G.J. Multiple protein sequence alignment from tertiary structure comparison: Assignment of global and residue confidence levels. Proteins 1992, 14, 309–323. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felsenstein, J. PHYLIP-Phylogeny Inference Package, Version 3.2. Cladistics 1989, 5, 164–166. [Google Scholar]
- Ropelewski, A.J.; Nicholas, H.B.; Gonzalez Mendez, R.R. MPI-PHYLIP: Parallelizing computationally intensive phylogenetic analysis routines for the analysis of large protein families. PLoS ONE 2010, 5, e13999. [Google Scholar] [CrossRef] [PubMed]
- Rambault, A. FigTree, v.1.4.4; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2010; Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 30 August 2022).
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’Neill, K.; Rob-bertse, B.; et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Barrett, T.; Benson, D.A.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; DiCuccio, M.; Edgar, R.; Federhen, S.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009, 37, D5–D15. [Google Scholar] [CrossRef] [Green Version]
- Maddison, D.R.; Schulz, K.-S.; Maddison, W.P. The Tree of Life Web Project. Zootaxa 2007, 1668, 19–40. [Google Scholar] [CrossRef]
- Durand, D.; Halldórsson, B.V.; Vernot, B. A hybrid micro-macroevolutionary approach to gene tree reconstruction. J. Comput. Biol. 2006, 13, 320–335. [Google Scholar] [CrossRef]
- Vernot, B.; Stolzer, M.; Goldman, A.; Durand, D. Reconciliation with non-binary species trees. J. Comput. Biol. 2008, 15, 981–1006. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Durand, D.; Farach-Colton, M. NOTUNG: A program for dating gene duplications and optimizing gene family trees. J. Comput. Biol. 2000, 7, 429–447. [Google Scholar] [CrossRef]
- Felsenstein, J. Inferring Phylogenies, 2nd ed.; Sinauer Associates, Inc.: Sunderland, MA, USA, 2003. [Google Scholar]
- Zmasek, C.M.; Eddy, S.R. ATV: Display and manipulation of annotated phylogenetic trees. Bioinformatics 2001, 17, 383–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelé, J.; Abdi, H.; Moreau, M.; Thybert, D.; Chabbert, M. Multidimensional scaling reveals the main evolutionary pathways of class A G-protein-coupled receptors. PLoS ONE 2011, 6, e19094. [Google Scholar] [CrossRef] [PubMed]
- Pelé, J.; Bécu, J.M.; Abdi, H.; Chabbert, M. Bios2mds: An R package for comparing orthologous protein families by metric multidimensional scaling. BMC Bioinform. 2012, 13, 133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moretti, S.; Armougom, F.; Wallace, I.M.; Higgins, D.G.; Jongeneel, C.V.; Notredame, C. The M-Coffee web server: A meta-method for computing multiple sequence alignments by combining alternative alignment methods. Nucleic Acids Res. 2007, 35, W645–W648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar]
- Furuta, H.; Kajita, A. Dimeric hemoglobin of the bivalve mollusc Anadara broughtonii: Complete amino acid sequence of the globin chain. Biochemistry 1983, 22, 917–922. [Google Scholar] [CrossRef]
- Ptitsyn, O.B.; Ting, K.L. Non-functional conserved residues in globins and their possible role as a folding nucleus. J. Mol. Biol. 1999, 291, 671–682. [Google Scholar] [CrossRef] [Green Version]
- Liong, E.C.; Dou, Y.; Scott, E.E.; Olson, J.S.; Phillips, G.N. Waterproofing the heme pocket. Role of proximal amino acid side chains in preventing hemin loss from myoglobin. J. Biol. Chem. 2001, 276, 9093–9100. [Google Scholar] [CrossRef] [Green Version]
- Kraus, D.W. Heme Proteins in Sulfide-Oxidizing Bacteria/Mollusc Symbioses. Am. Zool. 1995, 35, 112–120. [Google Scholar] [CrossRef]
- Suzuki, T.; Takagi, T.; Ohta, S. Amino acid sequence of the dimeric hemoglobin (Hb I) from the deep-sea cold-seep clam Calyptogena soyoae and the phylogenetic relationship with other molluscan globins. Biochim. Biophys. Acta 1989, 999, 254–259. [Google Scholar] [CrossRef]
- Suzuki, T.; Takagi, T.; Ohta, S. Primary structure of a dimeric haemoglobin from the deep-sea cold-seep clam Calyptogena soyoae. Biochem. J. 1989, 260, 177–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Dover, C.L. The Ecology of Deep-Sea Hydrothermal Vents; Princeton University Press: Princeton, NJ, USA, 2000. [Google Scholar]
- Vetter, R.D.; Wells, M.E.; Kurtsman, A.L.; Somero, G.N. Sulfide Detoxification by the Hydrothermal Vent Crab Bythograea thermydron and Other Decapod Crustaceans. Physiol. Zool. 1987, 60, 121–137. [Google Scholar] [CrossRef]
- Suzuki, T.; Arita, T.; Kawasaki, Y. The cDNA-derived amino acid sequence of chain II of the heterodimeric hemoglobin from the blood clam Barbatia virescens. Zool. Sci. 1995, 12, 453–455. [Google Scholar] [CrossRef]
- Montes-Rodríguez, I.M.; Rivera, L.E.; López-Garriga, J.; Cadilla, C.L. Characterization and Expression of the Lucina pectinata Oxygen and Sulfide Binding Hemoglobin Genes. PLoS ONE 2016, 11, e0147977. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Kawamichi, H.; Ohtsuki, R.; Iwai, M.; Fujikura, K. Isolation and cDNA-derived amino acid sequences of hemoglobin and myoglobin from the deep-sea clam Calyptogena kaikoi. Biochim. Biophys. Acta 2000, 1478, 152–158. [Google Scholar] [CrossRef]
- Kawano, K.; Iwasaki, N.; Suzuki, T. Notable diversity in hemoglobin expression patterns among species of the deep-sea clam, Calyptogena. Cell. Mol. Life Sci. 2003, 60, 1952–1956. [Google Scholar] [CrossRef]
- Chiancone, E.; Vecchini, P.; Verzili, D.; Ascoli, F.; Antonini, E. Dimeric and tetrameric hemoglobins from the mollusc Scapharca inaequivalvis. Structural and functional properties. J. Mol. Biol. 1981, 152, 577–592. [Google Scholar] [CrossRef]
- Suzuki, T.; Kawasaki, Y.; Arita, T.; Nakamura, A. Two-domain haemoglobin of the blood clam Barbatia lima resulted from the recent gene duplication of the single-domain delta chain. Biochem. J. 1996, 313 Pt 2, 561–566. [Google Scholar] [CrossRef]
- Bao, Y.B.; Wang, Q.; Guo, X.M.; Lin, Z.H. Structure and immune expression analysis of hemoglobin genes from the blood clam Tegillarca granosa. Genet. Mol. Res. 2013, 12, 3110–3123. [Google Scholar] [CrossRef]
- Dewilde, S.; Ebner, B.; Vinck, E.; Gilany, K.; Hankeln, T.; Burmester, T.; Kreiling, J.; Reinisch, C.; Vanfleteren, J.R.; Kiger, L.; et al. The nerve hemoglobin of the bivalve mollusc Spisula solidissima: Molecular cloning, ligand binding studies, and phylogenetic analysis. J. Biol. Chem. 2006, 281, 5364–5372. [Google Scholar] [CrossRef] [Green Version]
- Dewilde, S.; Angelini, E.; Kiger, L.; Marden, M.C.; Beltramini, M.; Salvato, B.; Moens, L. Structure and function of the globin and globin gene from the Antarctic mollusc Yoldia eightsi. Biochem. J. 2003, 370 Pt 1, 245–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Springer, B.A.; Sligar, S.G.; Olson, J.S.; Phillips, G.N. Mechanisms of Ligand Recognition in Myoglobin. Chem. Rev. 1994, 94, 699–714. [Google Scholar] [CrossRef]
- Egeberg, K.D.; Springer, B.A.; Sligar, S.G.; Carver, T.E.; Rohlfs, R.J.; Olson, J.S. The role of Val68(E11) in ligand binding to sperm whale myoglobin. Site-directed mutagenesis of a synthetic gene. J. Biol. Chem. 1990, 265, 11788–11795. [Google Scholar] [CrossRef]
- Ramirez, E.; Cruz, A.; Rodriguez, D.; Uchima, L.; Pietri, R.; Santana, A.; López, G.E. Effects of active site mutations in haemoglobin I from Lucina pectinata: A molecular dynamic study. Mol. Simul. 2008, 34, 715–725. [Google Scholar] [CrossRef] [Green Version]
- Springer, B.A.; Egeberg, K.D.; Sligar, S.G.; Rohlfs, R.J.; Mathews, A.J.; Olson, J.S. Discrimination between oxygen and carbon monoxide and inhibition of autooxidation by myoglobin. Site-directed mutagenesis of the distal histidine. J. Biol. Chem. 1989, 264, 3057–3060. [Google Scholar] [CrossRef]
- Birukou, I.; Schweers, R.L.; Olson, J.S. Distal histidine stabilizes bound O2 and acts as a gate for ligand entry in both subunits of adult human hemoglobin. J. Biol. Chem. 2010, 285, 8840–8854. [Google Scholar] [CrossRef] [Green Version]
- Jensen, F.B. Comparative analysis of autoxidation of haemoglobin. J. Exp. Biol. 2001, 204 Pt 11, 2029–2033. [Google Scholar] [CrossRef]
- Pietri, R.; Román-Morales, E.; López-Garriga, J. Hydrogen sulfide and hemeproteins: Knowledge and mysteries. Antioxid. Redox Signal. 2011, 15, 393–404. [Google Scholar] [CrossRef] [Green Version]
- Román-Morales, E.; Pietri, R.; Ramos-Santana, B.; Vinogradov, S.N.; Lewis-Ballester, A.; López-Garriga, J. Structural determinants for the formation of sulfhemeprotein complexes. Biochem. Biophys. Res. Commun. 2010, 400, 489–492. [Google Scholar] [CrossRef] [Green Version]
- Arp, A.J.; Childress, J.J.; Vetter, R.D. The sulphide-binding protein in the blood of the vestimentiferan tube-worm, Riftia pachyptila, is the extracellular haemoglobin. J. Exp. Biol. 1987, 128, 139–158. [Google Scholar] [CrossRef]
- Flores, J.F.; Hourdez, S.M. The zinc-mediated sulfide-binding mechanism of hydrothermal vent tubeworm 400-kDa hemoglobin. Cah. Biol. Mar. 2006, 47, 371–377. [Google Scholar]
- Carlson, M.L.; Regan, R.; Elber, R.; Li, H.; Phillips, G.N.; Olson, J.S.; Gibson, Q.H. Nitric oxide recombination to double mutants of myoglobin: Role of ligand diffusion in a fluctuating heme pocket. Biochemistry 1994, 33, 10597–10606. [Google Scholar] [CrossRef]
- Pietri, R.; Granell, L.; Cruz, A.; De Jesús, W.; Lewis, A.; Leon, R.; Garriga, J.L. Tyrosine B10 and heme-ligand interactions of Lucina pectinata hemoglobin II: Control of heme reactivity. Biochim. Biophys. Acta 2005, 1747, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Hockenhull-Johnson, J.D.; Stern, M.S.; Martin, P.; Dass, C.; Desiderio, D.M.; Wittenberg, J.B.; Walz, D.A. The amino acid sequence of hemoglobin II from the symbiont-harboring clam Lucina pectinata. J. Protein Chem. 1991, 10, 609–622. [Google Scholar] [CrossRef] [PubMed]
- Hockenhull-Johnson, J.D.; Stern, M.S.; Wittenberg, J.B.; Vinogradov, S.N.; Kapp, O.H.; Walz, D.A. The amino acid sequence of hemoglobin III from the symbiont-harboring clam Lucina pectinata. J. Protein Chem. 1993, 12, 261–277. [Google Scholar] [CrossRef] [PubMed]
- Peterson, E.S.; Huang, S.; Wang, J.; Miller, L.M.; Vidugiris, G.; Kloek, A.P.; Friedman, J.M. A comparison of functional and structural consequences of the tyrosine B10 and glutamine E7 motifs in two invertebrate hemoglobins (Ascaris suum and Lucina pectinata). Biochemistry 1997, 36, 13110–13121. [Google Scholar] [CrossRef] [PubMed]
- Wittenberg, J.B.; Bolognesi, M.; Wittenberg, B.A.; Guertin, M. Truncated hemoglobins: A new family of hemoglobins widely distributed in bacteria, unicellular eukaryotes, and plants. J. Biol. Chem. 2002, 277, 871–874. [Google Scholar] [CrossRef] [Green Version]
- Igarashi, J.; Kobayashi, K.; Matsuoka, A. A hydrogen-bonding network formed by the B10-E7-E11 residues of a truncated hemoglobin from Tetrahymena pyriformis is critical for stability of bound oxygen and nitric oxide detoxification. J. Biol. Inorg. Chem. 2011, 16, 599–609. [Google Scholar] [CrossRef]
- Tarricone, C.; Galizzi, A.; Coda, A.; Ascenzi, P.; Bolognesi, M. Unusual structure of the oxygen-binding site in the dimeric bacterial hemoglobin from Vitreoscilla sp. Structure 1997, 5, 497–507. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, B.D.; Zhao, X.; Vyas, K.; La Mar, G.N.; Lile, R.A.; Brucker, E.A.; Phillips, G.N.; Olson, J.; Wittenberg, J.B. Solution and crystal structures of a sperm whale myoglobin triple mutant that mimics the sulfide-binding hemoglobin from Lucina pectinata. J. Biol. Chem. 1998, 273, 9517–9526. [Google Scholar] [CrossRef]







| Organism | Globin | Abbreviation | Amino Acids in Positions: | |||
|---|---|---|---|---|---|---|
| B9 | B10 | E7 | E11 | |||
| L. pectinata | Hemoglobin I | HbILp | Phe | Phe | Gln | Phe |
| L. pectinata | Hemoglobin II | HbIILp | Phe | Tyr | Gln | Phe |
| L. pectinata | Hemoglobin III | HbIIILp | Phe | Tyr | Gln | Phe |
| C. nautilei | Hemoglobin IV | HbIVCn | Phe | Tyr | Gln | Phe |
| C. nautilei | Hemoglobin III | HbIIICn | Phe | Tyr | Gln | Phe |
| C. kaikoi | Hemoglobin II | HbIICk | Phe | Tyr | Gln | Phe |
| C. tsubasa | Hemoglobin II | HbIICt | Phe | Tyr | Gln | Phe |
| C. soyoae | Hemoglobin II | HbIICs | Phe | Tyr | Gln | Phe |
| C. kaikoi | Hemoglobin I | HbICk | Phe | Tyr | Gln | Phe |
| C. soyoae | Hemoglobin I | HbICs | Phe | Tyr | Gln | Phe |
| C. tsubasa | Hemoglobin I | HbICt | Phe | Tyr | Gln | Phe |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montes-Rodríguez, I.M.; Cadilla, C.L.; López-Garriga, J.; González-Méndez, R. Bioinformatic Characterization and Molecular Evolution of the Lucina pectinata Hemoglobins. Genes 2022, 13, 2041. https://doi.org/10.3390/genes13112041
Montes-Rodríguez IM, Cadilla CL, López-Garriga J, González-Méndez R. Bioinformatic Characterization and Molecular Evolution of the Lucina pectinata Hemoglobins. Genes. 2022; 13(11):2041. https://doi.org/10.3390/genes13112041
Chicago/Turabian StyleMontes-Rodríguez, Ingrid M., Carmen L. Cadilla, Juan López-Garriga, and Ricardo González-Méndez. 2022. "Bioinformatic Characterization and Molecular Evolution of the Lucina pectinata Hemoglobins" Genes 13, no. 11: 2041. https://doi.org/10.3390/genes13112041