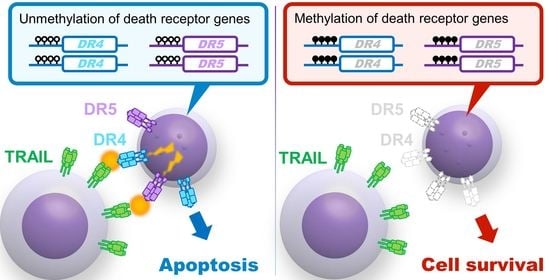
Epigenetic Modification of Death Receptor Genes for TRAIL and TRAIL Resistance in Childhood B-Cell Precursor Acute Lymphoblastic Leukemia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Leukemia Cell Lines and Patients’ Samples
2.2. 3H-Thymidine Uptake Assay
2.3. Cell-Surface Expression of DR4 and DR5
2.4. Real-Time Polymerase Chain Reaction Analysis
2.5. Bisulfite Sequencing
2.6. Gene Methylation and Gene Expression Analyses in Childhood BCP-ALL Cohort
2.7. Statistics
3. Results
3.1. Methylation Status of CpG Islands in the DR4 and DR5 Genes in BCP-ALL Cell Lines
3.2. Association of the Methylation Status of the DR4 and the DR5 with Their Gene and Cell-Surface Expressions and rhsTRAIL Sensitivity in BCP-ALL Cell Lines
3.3. Low Methylation Status of CpG Islands in the DR4 and DR5 Genes in ALL Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maude, S.L.; Laetsch, T.W.; Buechner, J.; Rives, S.; Boyer, M.; Bittencourt, H.; Bader, P.; Verneris, M.R.; Stefanski, H.E.; Myers, G.D.; et al. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N. Engl. J. Med. 2018, 378, 439–448. [Google Scholar] [CrossRef]
- Kantarjian, H.; Stein, A.; Gökbuget, N.; Fielding, A.K.; Schuh, A.C.; Ribera, J.M.; Wei, A.; Dombret, H.; Foà, R.; Bassan, R.; et al. Blinatumomab versus Chemotherapy for Advanced Acute Lymphoblastic Leukemia. N. Engl. J. Med. 2017, 376, 836–847. [Google Scholar] [CrossRef]
- Kolb, H.J.; Schmid, C.; Barrett, A.J.; Schendel, D.J. Graft-versus-leukemia reactions in allogeneic chimeras. Blood 2004, 103, 767–776. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.K.; McGuirk, J.P. Allogeneic Stem Cell Transplantation: A Historical and Scientific Overview. Cancer Res. 2016, 76, 6445–6451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickinson, A.M.; Norden, J.; Li, S.; Hromadnikova, I.; Schmid, C.; Schmetzer, H.; Jochem-Kolb, H. Graft-versus-Leukemia Effect Following Hematopoietic Stem Cell Transplantation for Leukemia. Front. Immunol. 2017, 8, 496. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Hayakawa, Y.; Smyth, M.J.; Kayagaki, N.; Yamaguchi, N.; Kakuta, S.; Iwakura, Y.; Yagita, H.; Okumura, K. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in surveillance of tumor metastasis by liver natural killer cells. Nat. Med. 2001, 7, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Smyth, M.J.; Cretney, E.; Takeda, K.; Wiltrout, R.H.; Sedger, L.M.; Kayagaki, N.; Yagita, H.; Okumura, K. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) contributes to interferon γ-dependent natural killer cell protection from tumor metastasis. J. Exp. Med. 2001, 193, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Schmaltz, C.; Alpdogan, O.; Kappel, B.J.; Muriglan, S.J.; Rotolo, J.A.; Ongchin, J.; Willis, L.M.; Greenberg, A.S.; Eng, J.M.; Crawford, J.M.; et al. T cells require TRAIL for optimal graft-versus-tumor activity. Nat. Med. 2002, 8, 1433–1437. [Google Scholar] [CrossRef]
- Lelaidier, M.; Dìaz-Rodriguez, Y.; Cordeau, M.; Cordeiro, P.; Haddad, E.; Herblot, S.; Duval, M. TRAIL-mediated killing of acute lymphoblastic leukemia by plasmacytoid dendritic cell-activated natural killer cells. Oncotarget 2015, 6, 29440–29455. [Google Scholar] [CrossRef] [Green Version]
- Pan, G.; O’Rourke, K.; Chinnaiyan, A.M.; Gentz, R.; Ebner, R.; Ni, J.; Dixit, V.M. The receptor for the cytotoxic ligand TRAIL. Science 1997, 276, 111–113. [Google Scholar] [CrossRef]
- Pan, G.; Ni, J.; Wei, Y.F.; Yu, G.; Gentz, R.; Dixit, V.M. An antagonist decoy receptor and death domain-containing receptor for TRAIL. Science 1997, 277, 815–818. [Google Scholar] [CrossRef]
- Aricò, M.; Valsecchi, M.G.; Camitta, B.; Schrappe, M.; Chessells, J.; Baruchel, A.; Gaynon, P.; Silverman, L.; Janka-Schaub, G.; Kamps, W.; et al. Outcome of treatment in children with Philadelphia chromosome-positive acute lymphoblastic leukemia. N. Engl. J. Med. 2000, 342, 998–1006. [Google Scholar] [CrossRef]
- Uno, K.; Inukai, T.; Kayagaki, N.; Goi, K.; Sato, H.; Nemoto, A.; Takahashi, K.; Kagami, K.; Yamaguchi, N.; Yagita, H.; et al. TNF-related apoptosis-inducing ligand (TRAIL) frequently induces apoptosis in Philadelphia chromosome-positive leukemia cells. Blood 2003, 101, 3658–3667. [Google Scholar] [CrossRef]
- Pui, C.H.; Gaynon, P.S.; Boyett, J.M.; Chessells, J.M.; Baruchel, A.; Kamps, W.; Silverman, L.B.; Biondi, A.; Harms, D.O.; Vilmer, E.; et al. Outcome of treatment in childhood acute lymphoblastic leukaemia with rearrangements of the 11q23 chromosomal region. Lancet 2002, 359, 1909–1915. [Google Scholar] [CrossRef]
- Inukai, T.; Zhang, X.; Goto, M.; Hirose, K.; Uno, K.; Akahane, K.; Nemoto, A.; Goi, K.; Sato, H.; Takahashi, K.; et al. Resistance of infant leukemia with MLL rearrangement to tumor necrosis factor-related apoptosis-inducing ligand: A possible mechanism for poor sensitivity to antitumor immunity. Leukemia 2006, 20, 2119–2129. [Google Scholar] [CrossRef] [Green Version]
- Saito, S.; Nakazawa, Y.; Sueki, A.; Matsuda, K.; Tanaka, M.; Yanagisawa, R.; Maeda, Y.; Sato, Y.; Okabe, S.; Inukai, T.; et al. Anti-leukemic potency of piggyBac-mediated CD19-specific T cells against refractory Philadelphia chromosome-positive acute lymphoblastic leukemia. Cytotherapy 2014, 16, 1257–1269. [Google Scholar] [CrossRef] [Green Version]
- Dufva, O.; Koski, J.; Maliniemi, P.; Ianevski, A.; Klievink, J.; Leitner, J.; Pölönen, P.; Hohtari, H.; Saeed, K.; Hannunen, T.; et al. Integrated drug profiling and CRISPR screening identify essential pathways for CAR T-cell cytotoxicity. Blood 2020, 135, 597–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akahane, K.; Inukai, T.; Zhang, X.; Hirose, K.; Kuroda, I.; Goi, K.; Honna, H.; Kagami, K.; Nakazawa, S.; Endo, K.; et al. Resistance of T-cell acute lymphoblastic leukemia to tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis. Exp. Hematol. 2010, 38, 885–895. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Inukai, T.; Hirose, K.; Akahane, K.; Kuroda, I.; Honna-Oshiro, H.; Kagami, K.; Goi, K.; Nakamura, K.; Kobayashi, M.; et al. Oncogenic fusion E2A-HLF sensitizes t(17;19)-positive acute lymphoblastic leukemia to TRAIL-mediated apoptosis by upregulating the expression of death receptors. Leukemia 2012, 26, 2483–2493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, M.; Miyake, K.; Kagami, K.; Abe, M.; Shinohara, T.; Watanabe, A.; Somazu, S.; Oshiro, H.; Goi, K.; Goto, H.; et al. Lack of association between deletion polymorphism of BIM gene and in vitro drug sensitivity in B-cell precursor acute lymphoblastic leukemia. Leuk. Res. 2017, 60, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Nordlund, J.; Bäcklin, C.L.; Wahlberg, P.; Busche, S.; Berglund, E.C.; Eloranta, M.L.; Flaegstad, T.; Forestier, E.; Frost, B.M.; Harila-Saari, A.; et al. Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol. 2013, 14, r105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paulsson, K.; Johansson, B. High hyperdiploid childhood acute lymphoblastic leukemia. Genes Chromosomes Cancer 2009, 48, 637–660. [Google Scholar] [CrossRef]
- Smiraglia, D.J.; Rush, L.J.; Frühwald, M.C.; Dai, Z.; Held, W.A.; Costello, J.F.; Lang, J.C.; Eng, C.; Li, B.; Wright, F.A.; et al. Excessive CpG island hypermethylation in cancer cell lines versus primary human malignancies. Hum. Mol. Genet. 2001, 10, 1413–1419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Topp, M.S.; Kufer, P.; Gökbuget, N.; Goebeler, M.; Klinger, M.; Neumann, S.; Horst, H.A.; Raff, T.; Viardot, A.; Schmid, M.; et al. Targeted therapy with the T-cell-engaging antibody blinatumomab of chemotherapy-refractory minimal residual disease in B-lineage acute lymphoblastic leukemia patients results in high response rate and prolonged leukemia-free survival. J. Clin. Oncol. 2011, 29, 2493–2498. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, A.; Inukai, T.; Kagami, K.; Abe, M.; Takagi, M.; Fukushima, T.; Fukushima, H.; Nanmoku, T.; Terui, K.; Ito, T.; et al. Resistance of t(17;19)-acute lymphoblastic leukemia cell lines to multiagents in induction therapy. Cancer Med. 2019, 8, 5274–5288. [Google Scholar] [CrossRef] [Green Version]
- Fischer, U.; Forster, M.; Rinaldi, A.; Risch, T.; Sungalee, S.; Warnatz, H.J.; Bornhauser, B.; Gombert, M.; Kratsch, C.; Stütz, A.M.; et al. Genomics and drug profiling of fatal TCF3-HLF-positive acute lymphoblastic leukemia identifies recurrent mutation patterns and therapeutic options. Nat. Genet. 2015, 47, 1020–1029. [Google Scholar] [CrossRef]
- Mouttet, B.; Vinti, L.; Ancliff, P.; Bodmer, N.; Brethon, B.; Cario, G.; Chen-Santel, C.; Elitzur, S.; Hazar, V.; Kunz, J.; et al. Durable remissions in TCF3-HLF positive acute lymphoblastic leukemia with blinatumomab and stem cell transplantation. Haematologica 2019, 104, e244–e247. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watanabe, A.; Miyake, K.; Akahane, K.; Goi, K.; Kagami, K.; Yagita, H.; Inukai, T. Epigenetic Modification of Death Receptor Genes for TRAIL and TRAIL Resistance in Childhood B-Cell Precursor Acute Lymphoblastic Leukemia. Genes 2021, 12, 864. https://doi.org/10.3390/genes12060864
Watanabe A, Miyake K, Akahane K, Goi K, Kagami K, Yagita H, Inukai T. Epigenetic Modification of Death Receptor Genes for TRAIL and TRAIL Resistance in Childhood B-Cell Precursor Acute Lymphoblastic Leukemia. Genes. 2021; 12(6):864. https://doi.org/10.3390/genes12060864
Chicago/Turabian StyleWatanabe, Atsushi, Kunio Miyake, Koshi Akahane, Kumiko Goi, Keiko Kagami, Hideo Yagita, and Takeshi Inukai. 2021. "Epigenetic Modification of Death Receptor Genes for TRAIL and TRAIL Resistance in Childhood B-Cell Precursor Acute Lymphoblastic Leukemia" Genes 12, no. 6: 864. https://doi.org/10.3390/genes12060864