Transcriptome-Based Identification of Genes Responding to the Organophosphate Pesticide Phosmet in Danio rerio
Abstract
:1. Introduction
2. Results
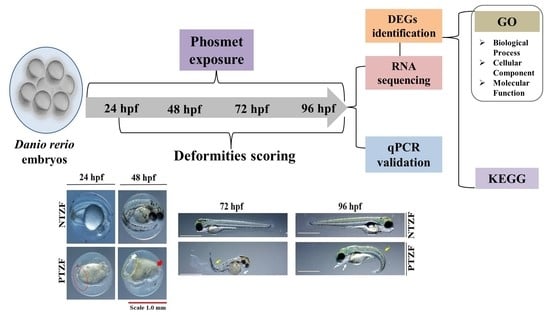
2.1. Effects of Phosmet on ZF Development
2.2. Transcriptome Analysis
2.3. Gene Ontology Term Enrichment
2.4. Functional Annotation of DEGs
2.5. qPCR Validation of Randomly Selected DEGs
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. ZF Maintenance and Embryo Collection
4.3. Pesticide Treatment and Deformities Scoring
4.4. Heartbeat Survey
4.5. TEER and VMER
4.6. Body Length Survey
4.7. RNA Sequencing Preparations
4.8. Transcriptome Alignment and Analysis of DEGs
4.9. Gene Ontology and Pathway Enrichment Analysis
4.10. Quantitative Real-Time PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mesnage, R.; Séralini, G.-E. Toxicity of pesticides on health and environment. Front. Public Health 2018, 6, 268. [Google Scholar] [CrossRef]
- Delfino, R.T.; Ribeiro, T.S.; Figueroa-Villar, J.D. Organophosphorus compounds as chemical warfare agents: A review. J. Braz. Chem. Soc. 2009, 20, 407–428. [Google Scholar] [CrossRef]
- Van der Veen, I.; de Boer, J. Phosphorus flame retardants: Properties, production, environmental occurrence, toxicity and analysis. Chemosphere 2012, 88, 1119–1153. [Google Scholar] [CrossRef]
- Ghimire, R.; Utyasheva, L.; Pokhrel, M.; Rai, N.; Chaudhary, B.; Prasad, P.N.; Bajracharya, S.R.; Basnet, B.; Das, K.D.; Pathak, N.K. Intentional pesticide poisoning and pesticide suicides in Nepal. Clin. Toxicol. 2021, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Karunarathne, A.; Bhalla, A.; Sethi, A.; Perera, U.; Eddleston, M. Importance of pesticides for lethal poisoning in India during 1999 to 2018: A systematic review. BMC Public Health 2021, 21, 1441. [Google Scholar] [CrossRef]
- Hertz-Picciotto, I.; Sass, J.B.; Engel, S.; Bennett, D.H.; Bradman, A.; Eskenazi, B.; Lanphear, B.; Whyatt, R. Organophosphate exposures during pregnancy and child neurodevelopment: Recommendations for essential policy reforms. PLoS Med. 2018, 15, e1002671. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Quezada, M.T.; Lucero, B.A.; Barr, D.B.; Steenland, K.; Levy, K.; Ryan, P.B.; Iglesias, V.; Alvarado, S.; Concha, C.; Rojas, E. Neurodevelopmental effects in children associated with exposure to organophosphate pesticides: A systematic review. Neurotoxicology 2013, 39, 158–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcês, A.; Pires, I.; Rodrigues, P. Teratological effects of pesticides in vertebrates: A review. J. Environ. Sci. Health Part B 2020, 55, 75–89. [Google Scholar] [CrossRef]
- Directorate of Plant Protection, Quarantine and Storage, India. Pesticide Wise Consumption of Indigenous Pesticides. 2021. Available online: http://ppqs.gov.in/sites/default/files/3.pesticidewise_consumption_of_indiginous_pesticides.xlsx (accessed on 19 October 2021).
- US EPA. 2006. Reregistration Eligibility Decision, Phosmet. Available online: https://www3.epa.gov/pesticides/chem_search/reg_actions/reregistration/red_PC-059201_31-Jul-06.pdf (accessed on 21 June 2021).
- Pesticide Info Data Base. California Pesticide Use, 2000–2018. Available online: https://www.pesticideinfo.org/california-search?title=Phosmet,All,All&chk=335&sk=all&cok=all&minyr=2000&maxyr=2018 (accessed on 21 October 2021).
- Bleyl, D. Embryotoxicity and teratogenicity of phosmet in mice. Arch. Fur Exp. Vet. 1980, 34, 791–795. [Google Scholar]
- US EPA, 2010. Risks of Phosmet Use to the Federally Threatened and Endangered California Tiger Salamander (Ambystoma californiense). Available online: https://www3.epa.gov/pesticides/endanger/litstatus/effects/redleg-frog/2010/phosmet/assessment.pdf (accessed on 21 October 2021).
- Lammer, E.; Carr, G.; Wendler, K.; Rawlings, J.; Belanger, S.; Braunbeck, T. Is the fish embryo toxicity test (FET) with the zebrafish (Danio rerio) a potential alternative for the fish acute toxicity test? Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2009, 149, 196–209. [Google Scholar] [CrossRef] [PubMed]
- Nagel, R. DarT: The embryo test with the Zebrafish Danio rerio—A general model in ecotoxicology and toxicology. Altex 2002, 19, 38–48. [Google Scholar]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [Green Version]
- Sandahl, J.F.; Baldwin, D.H.; Jenkins, J.J.; Scholz, N.L. Comparative thresholds for acetylcholinesterase inhibition and behavioral impairment in coho salmon exposed to chlorpyrifos. Environ. Toxicol. Chem. Int. J. 2005, 24, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Tierney, K.B.; Sampson, J.L.; Ross, P.S.; Sekela, M.A.; Kennedy, C.J. Salmon olfaction is impaired by an environmentally realistic pesticide mixture. Environ. Sci. Technol. 2008, 42, 4996–5001. [Google Scholar] [CrossRef]
- Vasamsetti, B.M.K.; Kim, N.S.; Chon, K.; Park, H.-H. Developmental Toxic Effects of Phosmet on Zebrafish (Danio rerio) Embryos. Korean J. Pestic. Sci. 2020, 24, 343–351. [Google Scholar] [CrossRef]
- Rahman, M.S.; Islam, S.M.; Haque, A.; Shahjahan, M. Toxicity of the organophosphate insecticide sumithion to embryo and larvae of zebrafish. Toxicol. Rep. 2020, 7, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Şişman, T. Dichlorvos-induced developmental toxicity in zebrafish. Toxicol. Ind. Health 2010, 26, 567–573. [Google Scholar] [CrossRef]
- Qayoom, I.; Balkhi, M.; Mukhtar, M.; Bhat, F.A.; Shah, F.A. Biochemical toxicity of organophosphate compounds in fishes. SKUAST J. Res. 2014, 16, 1–13. [Google Scholar]
- Laetz, C.A.; Baldwin, D.H.; Scholz, N.L. Sublethal neurotoxicity of organophosphate insecticides to juvenile coho salmon. Aquat. Toxicol. 2020, 221, 105424. [Google Scholar] [CrossRef]
- Schmitt, C.; McManus, M.; Kumar, N.; Awoyemi, O.; Crago, J. Comparative analyses of the neurobehavioral, molecular, and enzymatic effects of organophosphates on embryo-larval zebrafish (Danio rerio). Neurotoxicol. Teratol. 2019, 73, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Faria, M.; Garcia-Reyero, N.; Padrós, F.; Babin, P.J.; Sebastián, D.; Cachot, J.; Prats, E.; Arick, M.; Rial, E.; Knoll-Gellida, A. Zebrafish models for human acute organophosphorus poisoning. Sci. Rep. 2015, 5, 15591. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Canela, C.; Prats, E.; Piña, B.; Tauler, R. Assessment of chlorpyrifos toxic effects in zebrafish (Danio rerio) metabolism. Environ. Pollut. 2017, 220, 1231–1243. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Liu, X.; Yang, F.; Li, R.; Xiong, Y.; Fu, C.; Li, G.; Liu, S.; Zheng, C. Single and joint toxic effects of four antibiotics on some metabolic pathways of zebrafish (Danio rerio) larvae. Sci. Total. Environ. 2020, 716, 137062. [Google Scholar] [CrossRef] [PubMed]
- Ranjani, T.S.; Pitchika, G.K.; Yedukondalu, K.; Gunavathi, Y.; Daveedu, T.; Sainath, S.; Philip, G.; Pradeepkiran, J.A. Phenotypic and transcriptomic changes in zebrafish (Danio rerio) embryos/larvae following cypermethrin exposure. Chemosphere 2020, 249, 126148. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Lu, J.; Zhao, D. Toxicity and transcriptome sequencing (RNA-seq) analyses of adult zebrafish in response to exposure carboxymethyl cellulose stabilized iron sulfide nanoparticles. Sci. Rep. 2018, 8, 8083. [Google Scholar] [CrossRef] [PubMed]
- Lushchak, V.I.; Matviishyn, T.M.; Husak, V.V.; Storey, J.M.; Storey, K.B. Pesticide toxicity: A mechanistic approach. EXCLI J. 2018, 17, 1101. [Google Scholar]
- Park, M.Y.; Krishna Vasamsetti, B.M.; Kim, W.S.; Kang, H.J.; Kim, D.-Y.; Lim, B.; Cho, K.; Kim, J.S.; Chee, H.K.; Park, J.H. Comprehensive Analysis of Cardiac Xeno-Graft Unveils Rejection Mechanisms. Int. J. Mol. Sci. 2021, 22, 751. [Google Scholar]
- Akter, R.; Pervin, M.A.; Jahan, H.; Rakhi, S.F.; Reza, A.M.; Hossain, Z. Toxic effects of an organophosphate pesticide, envoy 50 SC on the histopathological, hematological, and brain acetylcholinesterase activities in stinging catfish (Heteropneustes fossilis). J. Basic Appl. Zool. 2020, 81, 47. [Google Scholar] [CrossRef]
- Rodríguez-Fuentes, G.; Rubio-Escalante, F.J.; Noreña-Barroso, E.; Escalante-Herrera, K.S.; Schlenk, D. Impacts of oxidative stress on acetylcholinesterase transcription, and activity in embryos of zebrafish (Danio rerio) following Chlorpyrifos exposure. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2015, 172, 19–25. [Google Scholar] [CrossRef]
- Richendrfer, H.; Creton, R. Chlorpyrifos and malathion have opposite effects on behaviors and brain size that are not correlated to changes in AChE activity. Neurotoxicology 2015, 49, 50–58. [Google Scholar] [CrossRef] [Green Version]
- Bui-Nguyen, T.M.; Baer, C.E.; Lewis, J.A.; Yang, D.; Lein, P.J.; Jackson, D.A. Dichlorvos exposure results in large scale disruption of energy metabolism in the liver of the zebrafish, Danio rerio. BMC Genom. 2015, 16, 853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boulekbache, H. Energy metabolism in fish development. Am. Zool. 1981, 21, 377–389. [Google Scholar] [CrossRef]
- Karami-Mohajeri, S.; Abdollahi, M. Mitochondrial dysfunction and organophosphorus compounds. Toxicol. Appl. Pharmacol. 2013, 270, 39–44. [Google Scholar] [CrossRef]
- Pearson, J.N.; Patel, M. The role of oxidative stress in organophosphate and nerve agent toxicity. Ann. N. Y. Acad. Sci. 2016, 1378, 17. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Avadhani, N.G. Cytochrome c oxidase dysfunction in oxidative stress. Free. Radic. Biol. Med. 2012, 53, 1252–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baden, K.N.; Murray, J.; Capaldi, R.A.; Guillemin, K. Early developmental pathology due to cytochrome c oxidase deficiency is revealed by a new zebrafish model. J. Biol. Chem. 2007, 282, 34839–34849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Giulio, R.T.; Hinton, D.E. The Toxicology of Fishes; CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar]
- Webb, S.E.; Miller, A.L. Calcium signalling during zebrafish embryonic development. Bioessays 2000, 22, 113–123. [Google Scholar] [CrossRef]
- Paudel, S.; Sindelar, R.; Saha, M. Calcium signaling in vertebrate development and its role in disease. Int. J. Mol. Sci. 2018, 19, 3390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Creton, R. The calcium pump of the endoplasmic reticulum plays a role in midline signaling during early zebrafish development. Dev. Brain Res. 2004, 151, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; Souders, C.L., II; Li, P.; Pang, S.; Qiu, L.; Martyniuk, C.J. Biological impacts of organophosphates chlorpyrifos and diazinon on development, mitochondrial bioenergetics, and locomotor activity in zebrafish (Danio rerio). Neurotoxicology Teratol. 2018, 70, 18–27. [Google Scholar] [CrossRef]
- Miura, G.I.; Yelon, D. A guide to analysis of cardiac phenotypes in the zebrafish embryo. Methods Cell Biol. 2011, 101, 161–180. [Google Scholar] [PubMed] [Green Version]
- Gut, P.; Reischauer, S.; Stainier, D.Y.; Arnaout, R. Little fish, big data: Zebrafish as a model for cardiovascular and metabolic disease. Physiol. Rev. 2017, 97, 889–938. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Lee, H.-C.; Fu, C.-Y.; Ding, Y.-Y.; Chen, J.-S.; Lee, M.-H.; Huang, W.-J.; Tsai, H.-J. MiR-1 and miR-206 target different genes to have opposing roles during angiogenesis in zebrafish embryos. Nat. Commun. 2013, 4, 2829. [Google Scholar] [CrossRef] [Green Version]
- Sehnert, A.J.; Huq, A.; Weinstein, B.M.; Walker, C.; Fishman, M.; Stainier, D.Y. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat. Genet. 2002, 31, 106–110. [Google Scholar] [CrossRef]
- Xu, X.; Meiler, S.E.; Zhong, T.P.; Mohideen, M.; Crossley, D.A.; Burggren, W.W.; Fishman, M.C. Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat. Genet. 2002, 30, 205–209. [Google Scholar] [CrossRef]
- Mably, J.D.; Burns, C.G.; Chen, J.-N.; Fishman, M.C.; Mohideen, M.-A.P. Heart of glass regulates the concentric growth of the heart in zebrafish. Curr. Biol. 2003, 13, 2138–2147. [Google Scholar] [CrossRef]
- Chen, Y.H.; Pai, C.W.; Huang, S.W.; Chang, S.N.; Lin, L.Y.; Chiang, F.T.; Lin, J.L.; Hwang, J.J.; Tsai, C.T. Inactivation of Myosin binding protein C homolog in zebrafish as a model for human cardiac hypertrophy and diastolic dysfunction. J. Am. Heart Assoc. 2013, 2, e000231. [Google Scholar] [CrossRef] [Green Version]
- Chopra, N.; Yang, T.; Asghari, P.; Moore, E.D.; Huke, S.; Akin, B.; Cattolica, R.A.; Perez, C.F.; Hlaing, T.; Knollmann-Ritschel, B.E. Ablation of triadin causes loss of cardiac Ca2+ release units, impaired excitation–contraction coupling, and cardiac arrhythmias. Proc. Natl. Acad. Sci. USA 2009, 106, 7636–7641. [Google Scholar] [CrossRef] [Green Version]
- Israeli-Rosenberg, S.; Manso, A.M.; Okada, H.; Ross, R.S. Integrins and integrin-associated proteins in the cardiac myocyte. Circ. Res. 2014, 114, 572–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, P.-Y.; Kutchukian, P.; Rajan Anand, D.; Imbriglio, J.; Andrews, C.; Padilla, H.; Vohra, A.; Lane, S.; Parker Jr, D.L.; Taracido, I.C. Cyp1 inhibition prevents doxorubicin-induced cardiomyopathy in a zebrafish heart failure model. Chembiochem A Eur. J. Chem. Biol. 2020, 21, 1905. [Google Scholar] [CrossRef] [PubMed]
- Créton, R.; Speksnijder, J.E.; Jaffe, L.F. Patterns of free calcium in zebrafish embryos. J. Cell Sci. 1998, 111, 1613–1622. [Google Scholar] [CrossRef]
- Paavola, J.; Schliffke, S.; Rossetti, S.; Kuo, I.Y.-T.; Yuan, S.; Sun, Z.; Harris, P.C.; Torres, V.E.; Ehrlich, B.E. Polycystin-2 mutations lead to impaired calcium cycling in the heart and predispose to dilated cardiomyopathy. J. Mol. Cell. Cardiol. 2013, 58, 199–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berardo, A.; Musumeci, O.; Toscano, A. Cardiological manifestations of mitochondrial respiratory chain disorders. Acta Myol. 2011, 30, 9. [Google Scholar]
- Pinho, B.R.; Santos, M.M.; Fonseca-Silva, A.; Valentão, P.; Andrade, P.B.; Oliveira, J.M. How mitochondrial dysfunction affects zebrafish development and cardiovascular function: An in vivo model for testing mitochondria-targeted drugs. Br. J. Pharmacol. 2013, 169, 1072–1090. [Google Scholar] [CrossRef] [Green Version]
- Jaga, K.; Dharmani, C. Ocular toxicity from pesticide exposure: A recent review. Environ. Health Prev. Med. 2006, 11, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Karanth, S.; Denovan-Wright, E.M.; Thisse, C.; Thisse, B.; Wright, J.M. The evolutionary relationship between the duplicated copies of the zebrafish fabp11 gene and the tetrapod FABP4, FABP5, FABP8 and FABP9 genes. FEBS J. 2008, 275, 3031–3040. [Google Scholar] [CrossRef]
- Zang, J.; Keim, J.; Kastenhuber, E.; Gesemann, M.; Neuhauss, S.C. Recoverin depletion accelerates cone photoresponse recovery. Open Biol. 2015, 5, 150086. [Google Scholar] [CrossRef] [Green Version]
- Zou, P.; Wu, S.-Y.; Koteiche, H.A.; Mishra, S.; Levic, D.S.; Knapik, E.; Chen, W.; Mchaourab, H.S. A conserved role of αA-crystallin in the development of the zebrafish embryonic lens. Exp. Eye Res. 2015, 138, 104–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farnsworth, D.R.; Posner, M.; Miller, A.C. Single cell transcriptomics of the developing zebrafish lens and identification of putative controllers of lens development. Exp. Eye Res. 2021, 206, 108535. [Google Scholar] [CrossRef]
- Inoue, M.; Miyahara, H.; Shiraishi, H.; Shimizu, N.; Tsumori, M.; Kiyota, K.; Maeda, M.; Umeda, R.; Ishitani, T.; Hanada, R. Leucyl-tRNA synthetase deficiency systemically induces excessive autophagy in zebrafish. Sci. Rep. 2021, 11, 8392. [Google Scholar] [CrossRef]
- Wang, Z.; Song, J.; Luo, L.; Ma, J. Loss of Leucyl-tRNA synthetase b leads to ILFS1-like symptoms in zebrafish. Biochem. Biophys. Res. Commun. 2018, 505, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Waldron, A.; Wilcox, C.; Francklyn, C.; Ebert, A. Knock-down of histidyl-tRNA synthetase causes cell cycle arrest and apoptosis of neuronal progenitor cells in vivo. Front. Cell Dev. Biol. 2019, 7, 67. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.J.; Collier, A.C.; Bowen, L.D.; Pritsos, K.L.; Goodrich, G.G.; Arger, K.; Cutter, G.; Pritsos, C.A. Polymorphisms in the NQO1, GSTT and GSTM genes are associated with coronary heart disease and biomarkers of oxidative stress. Mutat. Res./Genet. Toxicol. Environ. Mutagenes. 2009, 674, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Almannai, M.; Felemban, R.; Saleh, M.A.; Faqeih, E.A.; Alasmari, A.; AlHashem, A.; Mohamed, S.; Sunbul, R.; Al-Murshedi, F.; AlThihli, K. 6-Pyruvoyltetrahydropterin synthase deficiency: Review and report of 28 Arab subjects. Pediatric Neurol. 2019, 96, 40–47. [Google Scholar] [CrossRef] [Green Version]
- Lister, J.A. Larval but not adult xanthophore pigmentation in zebrafish requires GTP cyclohydrolase 2 (gch2) function. Pigment. Cell Melanoma Res. 2019, 32, 724–727. [Google Scholar] [CrossRef]
- Desvignes, T.; Fauvel, C.; Bobe, J. The nme gene family in zebrafish oogenesis and early development. Naunyn-Schmiedeberg’s Arch. Pharacol. 2011, 384, 439–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Huang, H.; Wu, Q. Characterization of the zebrafish Ugt repertoire reveals a new class of drug-metabolizing UDP glucuronosyltransferases. Mol. Pharmacol. 2014, 86, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Strähle, U.; Scholz, S.; Geisler, R.; Greiner, P.; Hollert, H.; Rastegar, S.; Schumacher, A.; Selderslaghs, I.; Weiss, C.; Witters, H. Zebrafish embryos as an alternative to animal experiments—A commentary on the definition of the onset of protected life stages in animal welfare regulations. Reprod. Toxicol. 2012, 33, 128–132. [Google Scholar] [CrossRef]
- Vsamsetti, B.M.K.; Kim, N.-S.; Chon, K.; Park, H.-H. Teratogenic and developmental toxic effects of etridiazole on zebrafish (Danio rerio) embryos. Appl. Biol. Chem. 2020, 63, 80. [Google Scholar] [CrossRef]






| Time | Deformity (%) | NTZF | PTZF |
|---|---|---|---|
| 24 hpf | Mortality | 0 | 17.50 ± 2.89 * |
| Edema symptoms | 2.50 ± 1.44 | 19.54 ± 7.11 * | |
| Abnormal somites | 2.50 ± 1.25 | 69.76 ± 8.02 * | |
| 48 hpf | Mortality | 0 | 32.50 ± 2.89 * |
| Low retina pigment | 0 | 63.07 ± 8.99 * | |
| Abnormal tail blood flow | 1.25 ± 1.25 | 53.85 ± 7.97 * | |
| Hyperemia | 0 | 44.22 ± 10.84 * | |
| 72 hpf | Mortality | 2.50 ± 1.44 | 43.75 ± 6.29 * |
| Unhatched embryos | 1.25 ± 1.25 | 78.22 ± 6.05 * | |
| Pericardial edema | 3.88 ± 1.30 | 61.63 ± 11.05 * | |
| Yolk sac edema | 2.57 ± 1.48 | 48.11 ± 9.31 * | |
| 96 hpf | Mortality | 2.50 ± 1.44 | 52.50 ± 3.23 * |
| Unhatched embryos | 0 | 82.29 ± 8.01 * | |
| Body curvature | 0 | 80.21 ± 5.15 * | |
| VMER | 25.00 ± 5.00 | 100.00 * | |
| TEER | 10.00 ± 5.77 | 90.00 ± 5.77 * |
| Samples | Total Reads | Clean Reads | Mapped Reads | Mapped Rate (%) | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|---|
| NTZF-1 | 72,783,012 | 71,783,716 | 60,458,452 | 84.22 | 98.91 | 96.05 | 43.62 |
| NTZF-2 | 61,925,392 | 60,857,682 | 51,896,692 | 85.28 | 98.91 | 96.06 | 44.95 |
| NTZF-3 | 60,878,020 | 59,726,314 | 51,603,485 | 86.4 | 98.85 | 95.87 | 46.00 |
| PTZF-1 | 69,410,088 | 68,273,094 | 45,753,478 | 67.02 | 98.89 | 96.11 | 53.24 |
| PTZF-2 | 66,557,046 | 65,493,004 | 55,524,220 | 84.78 | 98.92 | 96.13 | 49.44 |
| PTZF-3 | 74,253,266 | 73,041,798 | 61,232,206 | 83.83 | 98.97 | 96.27 | 47.69 |
| Term | Genes | p-Value | FDR |
|---|---|---|---|
| Metabolic pathways | gls2a, gls2b, gpx9, gda, hacd1, chia.2, gyg1b, tecrl2b, zgc:153896, elovl4b, zgc:153704, cyp7a1, zgc:103586, pgam2, pde6g, lpin1, ATP8, aoc1, LOC559107, ndufa4l2a, zgc:109982, pts, tyr, si:ch211-214p16.3, gch2, ampd1, mdh1ab, ND4L, tecrl2a, dhrs3a, atpv0e2, agxta, chs1, cyp21a2, aldoab, gal3st1a, ckmt2b, th2, si:dkey-78a14.5, dpm3, spam1, csgalnact1b, nat8l, ptges, atp5ia, nme2a, ND1, ND3, ND5, gpx8, ND2, si:dkey-78a14.4, aanat2, ndufa1, adssl1, si:ch211-217a12.1, ATP6, entpd8, alpi.2, tymp, ndufa4, elovl8a, entpd5a, zgc:112320, pigw, haao, CYTB, ckma, cel.1, gstk4, hao2, nme2b.2, pigh, pfkma, hpda, pnp4b, ND4, aldh3b2, smyd1a, b3gnt5a, mocs1, uox, st3gal7, si:ch211-106j24.1, pde6c, ndufa2, ugt5e1, fhit, sdhdb, acot19, agxtb, cyp2r1, COX3, COX1, COX2, tyrp1b, pde6d, acp5a *, sgpp2 *, galnt6 *, pik3c2g *, gal3st1b *, ugt5a1 *, atic *, glulc *, sqlea *, alp3 *, aldh3b1 *, p4ha1b *, paics *, elovl6l *, ethe1 *, plcd1a *, zgc:100864 *, zgc:154054 *, ugt1a5 *, cmasb *, aldob *, gstm.3 *, cyp24a1 *, ugt1a7 *, cbr1l *, zgc:123275 *, fut9d *, zgc:112332 *, b4galnt3b *, hao1 *, ugt1a1 *, mthfd1l *, si:ch73-337l15.2 *, ugt5a4 *, ptgs2b *, cmbl *, mgst1.2 *, rdh12l *, dao.1 *, zgc:163121 *, nansb *, si:ch211-276a23.5 *, cox4i1l *, pla2g4f.1 *, ugt1a2 *, ugt1a6 *, cyp3a65 *, cyp17a1 *, alas2 *, ptgs2a *, bhmt *, sqrdl *, nqo1 *, LOC565422 *, ptgis *, gstp1 *, ugt2a4 *, gstp2 * | 1.14402 × 10−53 | 1.61306 × 10−51 |
| Calcium signaling pathway | slc25a4, adrb2b, atp2a1l, mylk4b, stim2a, mylk4a, camk1gb, fgf4, ednraa, casq1a, trdn, tnnc2, p2rx5, avpr1aa, p2rx8, si:rp71-17i16.4, cacna1sb, casq2, calm1b, egf, ednrba, zgc:56235, casq1b, orai1a, si:dkey-247m21.3, fgf1a, p2rx3a, ryr3, atp2a1, phkg1b, tacr3a *, mcoln3a *, plcd1a *, p2rx1 *, si:dkey-251i10.1 *, cxcr4a *, ltb4r2a *, gna15.1 *, gna14 * | 2.39485 × 10−15 | 1.68837 × 10−13 |
| Phototransduction | opn1mw2, gngt1, opn1mw1, guca1c, gnat1, guca1d, pde6g, rhol, gnat2, rho, guca1e, rcvrnb, rcvrn2, grk7a, rcvrn3, calm1b, zgc:112320, cnga1 | 6.05641 × 10−15 | 2.84651 × 10−13 |
| Cardiac muscle contraction | atp2a1l, si:ch211-139a5.9, cacng6b, zgc:163073, zgc:86725, cacng1a, trdn, tnnt2d, atp1b1a, tpm4b, tnnt2e, cacna1sb, casq2, atp1b2b, CYTB, atp1a3b, atp1b4, smyhc1, atp2a1, atp1b2a, COX3, COX1, COX2, cox4i1l *, atp1a1a.2 * | 2.86244 × 10−14 | 1.00901 × 10−12 |
| Drug metabolism—other enzymes | zgc:103586, si:dkey-78a14.5, nme2a, si:dkey-78a14.4, gstk4, nme2b.2, aldh3b2, ugt5e1, ugt5a1 *, tpmt.1 *, tpmt.2 *, aldh3b1 *, ugt1a5 *, gstm.3 *, ugt1a7 *, ugt1a1 *, ugt5a4 *, mgst1.2 *, ugt1a2 *, ugt1a6 *, gstp1 *, ugt2a4 *, gstp2 * | 4.12652 × 10−13 | 1.16368 × 10−11 |
| Cell adhesion molecules | cldn7a, cldn19, mpz, cldn5b, mag, si:ch211-286o17.1, cldnj, cdh15, itgb1a, cd99l2, cldnf, cdh1, cldnc, cldn8, itgb1b.1, oclnb, zgc:110333, cldn7b, cldnb, oclna, cldn1, cldne, si:ch211-95j8.5, cldni, zgc:136892 | 2.61473 × 10−12 | 6.14462 × 10−11 |
| Aminoacyl-tRNA biosynthesis | trnS2, trnA, trnH, trnD, trnE, trnG, trnK, trnN, trnM, trnY, trnP, trnQ, trnC, trnR, trnI, trnL1, trnS1 | 4.82031 × 10−12 | 9.70947 × 10−11 |
| Biosynthesis of cofactors | pts, gch2, dhrs3a, nme2a, adssl1, alpi.2, haao, nme2b.2, hpda, mocs1, ugt5e1, ugt5a1, alp3, ugt1a5, ugt1a7, zgc:112332, ugt1a1, mthfd1l, ugt5a4, rdh12l, ugt1a2, ugt1a6, alas2, nqo1, ugt2a4 | 2.2858 × 10−11 | 3.55548 × 10−10 |
| Metabolism of xenobiotics by cytochrome P450 | gstk4, aldh3b2, ugt5e1, ugt5a1,aldh3b1, ugt1a5, gstm.3, ugt1a7, cbr1l, ugt1a1, ugt5a4, mgst1.2, ugt1a2, ugt1a6, gstp1, ugt2a4, gstp2 | 2.50462 × 10−11 | 3.55548 × 10−10 |
| Regulation of actin cytoskeleton | cfl2, cxcl12b, mylk4b, mylk4a, fgf4, mylpfb, cxcl12a, LOC101885790, si:dkey-44g17.6, itga10, egf, tmsb, scinla, pfn2l, itgb1a, fgf1a, si:ch73-116o1.2, brk1, itga2.2, itgb1b.1, cxcr4a, rac2, zgc:86896, myh9a, fn1b, itgb3a, gsnb, pfn1, cfl1l, zgc:101810 | 2.52162 ×10−11 | 3.55548 ×10−10 |
| Gene Symbol | Fold Change (RNA-Seq) | Fold Change (qPCR) (Mean ± SD) |
|---|---|---|
| gna14 | 6.13 | 4.17 ± 0.14 |
| fn1b | 4.14 | 2.13 ± 0.21 |
| gstp2 | 12.09 | 5.15 ± 1.28 |
| gls2b | −7.25 | −4.40 ± 0.56 |
| cnga1 | −2.07 | −2.84 ± 0.55 |
| adrb2b | −4.27 | −3.91 ± 1.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasamsetti, B.M.K.; Chon, K.; Kim, J.; Oh, J.-A.; Yoon, C.-Y.; Park, H.-H. Transcriptome-Based Identification of Genes Responding to the Organophosphate Pesticide Phosmet in Danio rerio. Genes 2021, 12, 1738. https://doi.org/10.3390/genes12111738
Vasamsetti BMK, Chon K, Kim J, Oh J-A, Yoon C-Y, Park H-H. Transcriptome-Based Identification of Genes Responding to the Organophosphate Pesticide Phosmet in Danio rerio. Genes. 2021; 12(11):1738. https://doi.org/10.3390/genes12111738
Chicago/Turabian StyleVasamsetti, Bala Murali Krishna, Kyongmi Chon, Juyeong Kim, Jin-A Oh, Chang-Young Yoon, and Hong-Hyun Park. 2021. "Transcriptome-Based Identification of Genes Responding to the Organophosphate Pesticide Phosmet in Danio rerio" Genes 12, no. 11: 1738. https://doi.org/10.3390/genes12111738