3′quant mRNA-Seq of Porcine Liver Reveals Alterations in UPR, Acute Phase Response, and Cholesterol and Bile Acid Metabolism in Response to Different Dietary Fats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Diets
2.2. RNA Isolation and 3′Quant mRNA Library Construction and Sequencing
2.3. Bioinformatic Analysis
2.4. Quantitative PCR
3. Results
3.1. Fatty Acids Profiles of Diets Used in the Experiment
3.2. 3′Quant mRNA Statistics and DEGs Identified in the Liver after Different Dietary Treatments
3.3. Functional Analysis of Identified DEGs with STRING
3.4. Functional Analysis of Identified DEGs with IPA
3.5. Identification of Hub Genes with Cytohubba
3.6. Validation of Quant 3′mRNA Profiling by qPCR
4. Discussion
4.1. Biosynthesis and Catabolism of Cholesterol are Inhibited in Animals Obtaining Beef Tallow in the Diet
4.2. Acute-Phase Response Signaling is Activated in the Beef Tallow Group when Compared to both Rapeseed Oil and Coconut Oil
4.3. Unfolded Protein Response is Activated in both the Beef Tallow and Coconut Oil Groups when Compared to the Rapeseed Oil Group
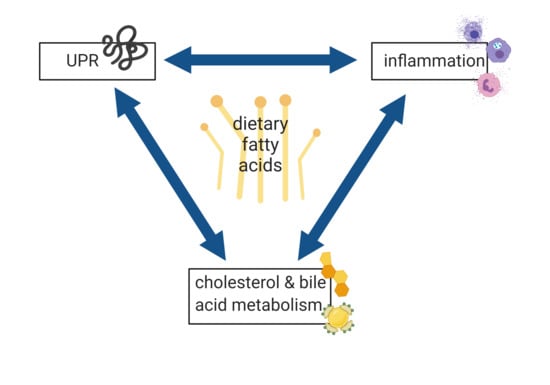
4.4. UPR, Inflammation, and Cholesterol and Bile Acid Metabolism are the Main Processes Affected by Dietary Fatty Acids
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Othman, R. Dietary lipids and cancer. Libyan J. Med. 2007, 2, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Zanoaga, O.; Jurj, A.; Raduly, L.; Cojocneanu-Petric, R.; Fuentes-Mattei, E.; Wu, O.; Braicu, C.; Gherman, C.D.; Berindan-Neagoe, I. Implications of dietary ω-3 and ω-6 polyunsaturated fatty acids in breast cancer. Exp. Ther. Med. 2018, 15, 1167–1176. [Google Scholar] [CrossRef] [PubMed]
- Halade, G.V.; Black, L.M.; Verma, M.K. Paradigm shift-Metabolic transformation of docosahexaenoic and eicosapentaenoic acids to bioactives exemplify the promise of fatty acid drug discovery. Biotechnol. Adv. 2018, 36, 935–953. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.L.; Carlson, D.F.; Largaespada, D.A.; Hackett, P.B.; Fahrenkrug, S.C. Engineered Swine Models of Cancer. Front Genet. 2016, 7, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cevallos, W.H.; Holmes, W.L.; Myers, R.N.; Smink, R.D. Swine in atherosclerosis research: Development of an experimental animal model and study of the effect of dietary fats on cholesterol metabolism. Atherosclerosis 1979, 34, 303–317. [Google Scholar] [CrossRef]
- Vitali, M.; DiMauro, C.; Sirri, R.; Zappaterra, M.; Zambonelli, P.; Manca, E.; Sami, D.; Fiego, D.P.L.; Davoli, R. Effect of dietary polyunsaturated fatty acid and antioxidant supplementation on the transcriptional level of genes involved in lipid and energy metabolism in swine. PLoS ONE 2018, 13, e0204869. [Google Scholar] [CrossRef] [PubMed]
- Szostak, A.; Ogłuszka, M.; Pas, M.F.W.T.; Poławska, E.; Urbański, P.; Juszczuk-Kubiak, E.; Blicharski, T.; Pareek, C.S.; Dunkelberger, J.R.; Horbańczuk, J.O.; et al. Effect of a diet enriched with omega-6 and omega-3 fatty acids on the pig liver transcriptome. Genes Nutr. 2016, 11, 9. [Google Scholar] [CrossRef] [Green Version]
- Ogłuszka, M.; Szostak, A.; Pas, M.F.T.; Poławska, E.; Urbański, P.; Blicharski, T.; Pareek, C.; Juszczuk-Kubiak, E.; Dunkelberger, J.R.; Horbańczuk, J.O.; et al. A porcine gluteus medius muscle genome-wide transcriptome analysis: Dietary effects of omega-6 and omega-3 fatty acids on biological mechanisms. Genes Nutr. 2017, 12, 4. [Google Scholar] [CrossRef] [Green Version]
- Oczkowicz, M.; Świątkiewicz, M.; Ropka-Molik, K.; Gurgul, A.; Żukowski, K. Effects of Different Sources of Fat in the Diet of Pigs on the Liver Transcriptome Estimated by RNA-Seq. Ann. Anim. Sci. 2016, 16, 1073–1090. [Google Scholar] [CrossRef] [Green Version]
- Świątkiewicz, M.; Oczkowicz, M.; Ropka-Molik, K.; Hanczakowska, E. The effect of dietary fatty acid composition on adipose tissue quality and expression of genes related to lipid metabolism in porcine livers. Anim. Feed Sci. Technol. 2016, 216, 204–215. [Google Scholar] [CrossRef]
- Oczkowicz, M.; Szmatoła, T.; Świątkiewicz, M.; Pawlina-Tyszko, K.; Gurgul, A.; Ząbek, T. Corn dried distillers grains with solubles (cDDGS) in the diet of pigs change the expression of adipose genes that are potential therapeutic targets in metabolic and cardiovascular diseases. BMC Genom. 2018, 19, 864. [Google Scholar] [CrossRef] [PubMed]
- Oczkowicz, M.; Szmatoła, T.; Świątkiewicz, M. Source of Dietary Fat in Pig Diet Affects Adipose Expression of Genes Related to Cancer, Cardiovascular, and Neurodegenerative Diseases. Genes 2019, 10, 948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef] [Green Version]
- Simopoulos, A.P. An Increase in the Omega-6/Omega-3 Fatty Acid Ratio Increases the Risk for Obesity. Nutrients 2016, 8, 128. [Google Scholar] [CrossRef] [Green Version]
- De Wit, N.; Derrien, M.; Bosch-Vermeulen, H.; Oosterink, E.; Keshtkar, S.; Duval, C.; Bosch, J.D.V.-V.D.; Kleerebezem, M.; Muller, M.; Van Der Meer, R. Saturated fat stimulates obesity and hepatic steatosis and affects gut microbiota composition by an enhanced overflow of dietary fat to the distal intestine. Am. J. Physiol. Gastrointest. Liver. Physiol. 2012, 303, G589–G599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alagawany, M.; Abd El-Hack, M.E.; Al-Sagheer, A.A.; Naiel, M.A.; Saadeldin, I.M.; Swelum, A.A. Dietary Cold Pressed Watercress and Coconut Oil Mixture Enhances Growth Performance, Intestinal Microbiota, Antioxidant Status, and Immunity of Growing Rabbits. Animals 2018, 8, 212. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.-M.; Wang, X.-H.; Hao, L.-H.; Wang, H.; Zhang, X.; Muhammad, I.; Qi, Y.; Li, G.-L.; Sun, X.-Q. Nrf2 is crucial for the down-regulation of Cyp7a1 induced by arachidonic acid in Hepg2 cells. Environ. Toxicol. Pharmacol. 2017, 52, 21–26. [Google Scholar] [CrossRef]
- Ranoa, D.R.; Kelley, S.L.; Tapping, R.I. Human lipopolysaccharide-binding protein (LBP) and CD14 independently deliver triacylated lipoproteins to Toll-like receptor 1 (TLR1) and TLR2 and enhance formation of the ternary signaling complex. J. Biol. Chem. 2013, 288, 9729–9741. [Google Scholar] [CrossRef] [Green Version]
- Bueno, A.A.; Oyama, L.M.; Motoyama, C.S.D.M.; Biz, C.R.D.S.; Silveira, V.L.; Ribeiro, E.B.; Nascimento, C.M.O.D. Long chain saturated fatty acids increase haptoglobin gene expression in C57BL/6J mice adipose tissue and 3T3-L1 cells. Eur. J. Nutr. 2010, 49, 235–241. [Google Scholar] [CrossRef]
- Yamada, S.; Kamada, N.; Amiya, T.; Nakamoto, N.; Nakaoka, T.; Kimura, M.; Saito, Y.; Ejima, C.; Kanai, T.; Saito, Y. Gut microbiota-mediated generation of saturated fatty acids elicits inflammation in the liver in murine high-fat diet-induced steatohepatitis. BMC Gastroenterol. 2017, 17, 136. [Google Scholar] [CrossRef]
- Nishimura, Y.; Moriyama, M.; Kawabe, K.; Satoh, H.; Takano, K.; Azuma, Y.-T.; Nakamura, Y. Lauric Acid Alleviates Neuroinflammatory Responses by Activated Microglia: Involvement of the GPR40-Dependent Pathway. Neurochem. Res. 2018, 43, 1723–1735. [Google Scholar] [CrossRef] [PubMed]
- Grootjans, J.; Kaser, A.; Kaufman, R.J.; Blumberg, R.S. The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 2016, 16, 469–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volmer, R.; van der Ploeg, K.; Ron, D. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc. Natl. Acad. Sci. USA 2013, 110, 4628–4633. [Google Scholar] [CrossRef] [Green Version]
- Volmer, R.; Ron, D. Lipid-dependent regulation of the unfolded protein response. Curr. Opin. Cell Biol. 2015, 33, 67–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Dror, K.; Birk, R. Oleic acid ameliorates palmitic acid-induced ER stress and inflammation markers in naive and cerulein-treated exocrine pancreas cells. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [Green Version]
- McDonnell, E.; Peterson, B.S.; Bomze, H.M.; Hirschey, M.D. SIRT3 regulates progression and development of diseases of aging. Trends Endocrinol. Metab. 2015, 26, 486–492. [Google Scholar] [CrossRef] [Green Version]
- Finley, L.W.; Carracedo, A.; Lee, J.J.; Souza, A.; Egia, A.; Zhang, J.; Teruya-Feldstein, J.; Moreira, P.; Cardoso, S.M.; Clish, C.B.; et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell. 2011, 19, 416–428. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Xin, T.; Li, D.; Wang, C.; Zhu, H.; Zhou, H. Therapeutic effect of Sirtuin 3 on ameliorating nonalcoholic fatty liver disease: The role of the ERK-CREB pathway and Bnip3-mediated mitophagy. Redox Biol. 2018, 18, 229–243. [Google Scholar] [CrossRef]
- Li, S.; Dou, X.; Ning, H.; Song, Q.; Wei, W.; Zhang, X.; Shen, C.; Li, J.; Sun, C.; Song, Z. Sirtuin 3 acts as a negative regulator of autophagy dictating hepatocyte susceptibility to lipotoxicity. Hepatology 2017, 66, 936–952. [Google Scholar] [CrossRef]
- Pizzinga, M.; Harvey, R.F.; Garland, G.D.; Mordue, R.; Dezi, V.; Ramakrishna, M.; Sfakianos, A.; Monti, M.; Mulroney, T.E.; Poyry, T.; et al. The cell stress response: Extreme times call for post-transcriptional measures. Wiley Interdiscip. Rev. RNA 2020, 11, e1578. [Google Scholar] [CrossRef]
- Xing, Y.-H.; Yao, R.-W.; Zhang, Y.; Guo, C.-J.; Jiang, S.; Xu, G.; Dong, R.; Yang, L.; Chen, L.-L. SLERT Regulates DDX21 Rings Associated with Pol I Transcription. Cell 2017, 169, 664–678.e16. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Zhang, Y.; Ma, L.; Cheng, Q.; Li, G.; Tong, T. Enhanced NOLC1 promotes cell senescence and represses hepatocellular carcinoma cell proliferation by disturbing the organization of nucleolus. Aging Cell. 2017, 16, 726–737. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.D.; Collins, J.C.; Korona, B.; Ghalei, H.; Karbstein, K. A kinase-dependent checkpoint prevents escape of immature ribosomes into the translating pool. PLoS Biol. 2019, 17, e3000329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, B.; Wei, C.; Qiao, Z.; Han, W.; Chai, X.; Lu, J.; Gao, C.; Dong, R.; Gao, D.; Huang, C.; et al. eIF3a mediates HIF1α-dependent glycolytic metabolism in hepatocellular carcinoma cells through translational regulation. Am. J. Cancer Res. 2019, 9, 1079–1090. [Google Scholar]
- Cao, S.S.; Luo, K.L.; Shi, L. Endoplasmic Reticulum Stress Interacts With Inflammation in Human Diseases. J. Cell Physiol. 2016, 231, 288–294. [Google Scholar] [CrossRef]
- Allen, K.; Jaeschke, H.; Copple, B.L. Bile acids induce inflammatory genes in hepatocytes: A novel mechanism of inflammation during obstructive cholestasis. Am. J. Pathol. 2011, 178, 175–186. [Google Scholar] [CrossRef]
- Libert, C.; Brouckaert, P.; Fiers, W. Protection by α 1-acid glycoprotein against tumor necrosis factor-induced lethality. J. Exp. Med. 1994, 180, 1571–1575. [Google Scholar] [CrossRef]
- Cai, L.; Oyeniran, C.; Biswas, D.D.; Allegood, J.; Milstien, S.; Kordula, T.; Maceyka, M.; Spiegel, S. ORMDL proteins regulate ceramide levels during sterile inflammation. J. Lipid Res. 2016, 57, 1412–1422. [Google Scholar] [CrossRef] [Green Version]
- Porez, G.; Gross, B.; Prawitt, J.; Gheeraert, C.; Berrabah, W.; Alexandre, J.; Staels, B.; Lefebvre, P. The hepatic orosomucoid/α1-acid glycoprotein gene cluster is regulated by the nuclear bile acid receptor FXR. Endocrinology 2013, 154, 3690–3701. [Google Scholar] [CrossRef] [Green Version]
- Khovidhunkit, W.; Kim, M.-S.; Memon, R.A.; Shigenaga, J.K.; Moser, A.H.; Feingold, K.R.; Grunfeld, C. Effects of infection and inflammation on lipid and lipoprotein metabolism: Mechanisms and consequences to the host. J. Lipid Res. 2004, 45, 1169–1196. [Google Scholar] [CrossRef] [Green Version]
- Feingold, K.R.; Spady, D.K.; Pollock, A.S.; Moser, A.H.; Grunfeld, C. Endotoxin, TNF, and IL-1 decrease cholesterol 7 α-hydroxylase mRNA levels and activity. J. Lipid Res. 1996, 37, 223–228. [Google Scholar] [PubMed]
- Han, S.; Lone, M.A.; Schneiter, R.; Chang, A. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. USA 2010, 107, 5851–5856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, M.K.; Wallington-Beddoe, C.T.; Pitson, S.M. Sphingolipids and the unfolded protein response. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 1483–1494. [Google Scholar] [CrossRef] [PubMed]
- Reverendo, M.; Mendes, A.; Argüello, R.J.; Gatti, E.; Pierre, P. At the crossway of ER-stress and proinflammatory responses. FEBS J. 2019, 286, 297–310. [Google Scholar] [CrossRef] [Green Version]
- Bashiri, A.; Nesan, D.; Tavallaee, G.; Sue-Chue-Lam, I.; Chien, K.; Maguire, G.F.; Naples, M.; Zhang, J.; Magomedova, L.; Adeli, K.; et al. Cellular cholesterol accumulation modulates high fat high sucrose (HFHS) diet-induced ER stress and hepatic inflammasome activation in the development of non-alcoholic steatohepatitis. Biochim. Biophys. Acta 2016, 1861, 594–605. [Google Scholar] [CrossRef]
- Widenmaier, S.B.; Snyder, N.A.; Nguyen, T.B.; Arduini, A.; Lee, G.Y.; Arruda, A.P.; Saksi, J.; Bartelt, A.; Hotamisligil, G.S. NRF1 Is an ER Membrane Sensor that Is Central to Cholesterol Homeostasis. Cell 2017, 171, 1094–1109.e15. [Google Scholar] [CrossRef]
- Silva-Islas, C.A.; Maldonado, P.D. Canonical and non-canonical mechanisms of Nrf2 activation. Pharmacol. Res. 2018, 134, 92–99. [Google Scholar] [CrossRef]
- Lee, M.-H.; Appleton, K.M.; El-Shewy, H.M.; Sorci-Thomas, M.G.; Thomas, M.J.; Lopes-Virella, M.F.; Luttrell, L.M.; Hammad, S.M.; Klein, R.L. S1P in HDL promotes interaction between SR-BI and S1PR1 and activates S1PR1-mediated biological functions: Calcium flux and S1PR1 internalization. J. Lipid Res. 2017, 58, 325–338. [Google Scholar] [CrossRef] [Green Version]
- Motiani, R.K.; Abdullaev, I.F.; Trebak, M. A novel native store-operated calcium channel encoded by Orai3: Selective requirement of Orai3 versus Orai1 in estrogen receptor-positive versus estrogen receptor-negative breast cancer cells. J. Biol. Chem. 2010, 285, 19173–19183. [Google Scholar] [CrossRef] [Green Version]









| Sample Name | No of Raw Reads | No of Mapped Reads | Percentage of Mapped Reads (%) |
|---|---|---|---|
| 17w | 3,355,869 | 2,635,495 | 78.5 |
| 19w | 2,460,130 | 1,927,026 | 78.3 |
| 20w | 2,029,705 | 1,615,350 | 79.6 |
| 21w | 2,819,316 | 2,187,454 | 77.6 |
| 22w | 2,657,611 | 2,111,384 | 79.4 |
| 24w | 2,298,911 | 1,803,582 | 78.5 |
| 41w | 3,469,934 | 2,760,471 | 79.6 |
| 42w | 2,223,698 | 1,817,882 | 81.8 |
| 43w | 4,124,251 | 3,197,675 | 77.5 |
| 45w | 2,157,086 | 1,710,536 | 79.3 |
| 46w | 2,415,641 | 1,932,621 | 80.0 |
| 48w | 2,470,229 | 1,981,470 | 80.2 |
| 49w | 2,715,605 | 2,203,190 | 81.1 |
| 52w | 2,631,687 | 2,119,487 | 80.5 |
| 53w | 2,110,920 | 1,648,309 | 78.1 |
| 54w | 2,031,614 | 1,625,581 | 80.0 |
| 55w | 4,000,000 | 3,266,253 | 81.7 |
| 56w | 4,000,000 | 3,262,080 | 81.6 |
| Ensembl ID | Base Mean | log2 Fold Change | p-Adjusted | Gene Description | Gene Symbol |
|---|---|---|---|---|---|
| Beef Tallow vs. Coconut Oil | |||||
| ENSSSCG00000021443 | 109 | −1.46 | 0.0005 | serum/glucocorticoid regulated kinase 1 | SGK1 |
| ENSSSCG00000032381 | 221 | −1.42 | 0.0000 | lipopolysaccharide induced TNF factor | LITAF |
| ENSSSCG00000004700 | 288 | −1.34 | 0.0007 | protein disulfide isomerase family A member 3 | PDIA3 |
| ENSSSCG00000028758 | 177 | −1.28 | 0.0000 | lipopolysaccharide binding protein | LBP |
| ENSSSCG00000000892 | 127 | −1.23 | 0.0008 | histidine ammonia-lyase | HAL |
| ENSSSCG00000033214 | 460 | −1.16 | 0.0098 | glycine N-methyltransferase | GNMT |
| ENSSSCG00000016174 | 361 | −1.16 | 0.0185 | fibronectin 1 | FN1 |
| ENSSSCG00000008998 | 23136 | −1.01 | 0.0001 | fibrinogen α chain | FGA |
| ENSSSCG00000013514 | 256 | −0.94 | 0.0001 | LRRCT domain-containing protein | PLIN5 |
| ENSSSCG00000008550 | 317 | −0.86 | 0.0124 | solute carrier family 5 member 6 | SLC5A6 |
| ENSSSCG00000022126 | 122 | −0.79 | 0.0025 | epidermal growth factor receptor | EGFR |
| ENSSSCG00000002487 | 1830 | −0.78 | 0.0027 | α-1-antichymotrypsin 2 | SERPINA3-2 |
| ENSSSCG00000002749 | 37957 | −0.59 | 0.0162 | haptoglobin | HP |
| ENSSSCG00000011453 | 1755 | −1.43 | 0.0000 | inter-α-trypsin inhibitor heavy chain 4 | ITIH4 |
| ENSSSCG00000011741 | 340 | 0.45 | 0.0185 | golgi integral membrane protein 4 | GOLIM4 |
| ENSSSCG00000027072 | 172 | 0.56 | 0.0112 | ATP synthase inhibitory factor subunit 1 | ATP5IF1 |
| ENSSSCG00000002529 | 355 | 0.65 | 0.0036 | 40S ribosomal protein S21 | RPS21 |
| ENSSSCG00000008829 | 182 | 0.80 | 0.0076 | OCIA domain containing 2 | OCIAD2 |
| ENSSSCG00000026044 | 221 | 1.34 | 0.0005 | farnesyl-diphosphate farnesyltransferase 1 | FDFT1 |
| ENSSSCG00000006238 | 84 | 2.11 | 0.0007 | cytochrome P450 family 7 subfamily A member 1 | CYP7A1 |
| Rapeseed Oil vs. Beef Tallow | |||||
| ENSSSCG00000006238 | 100 | −2.63 | 4 × 10-8 | cytochrome P450 family 7 subfamily A member 1 | CYP7A1 |
| ENSSSCG00000028821 | 27 | −2.14 | 1.6 × 105 | SAS-6 centriolar assembly protein | SASS6 |
| ENSSSCG00000004586 | 19 | −2.02 | 6.5 × 105 | family with sequence similarity 81 member A | FAM81A |
| ENSSSCG00000033822 | 110 | −1.91 | 6.6 × 105 | thyroid hormone responsive | THRSP |
| ENSSSCG00000026044 | 252 | −1.80 | 1.6 × 105 | farnesyl-diphosphate farnesyltransferase 1 | FDFT1 |
| ENSSSCG00000006719 | 94 | −1.41 | 1.6 × 107 | hydroxy-delta-5-steroid dehydrogenase, 3 β- and steroid delta-isomerase 1 | HSD3B1 |
| ENSSSCG00000006040 | 186 | −1.20 | 2.6 × 106 | dihydropyrimidinase | DPYS |
| ENSSSCG00000001849 | 153 | −0.89 | 1.7 × 104 | alanyl aminopeptidase, membrane | ANPEP |
| ENSSSCG00000039388 | 448 | −0.84 | 8.3 × 105 | ||
| ENSSSCG00000023686 | 5283 | −0.78 | 3.4 × 104 | transthyretin | TTR |
| ENSSSCG00000015106 | 124 | 1.23 | 1.5 × 104 | hypoxia up-regulated 1 | HYOU1 |
| ENSSSCG00000032381 | 218 | 1.35 | 3.4 × 104 | lipopolysaccharide induced TNF factor | LITAF |
| ENSSSCG00000011297 | 222 | 1.36 | 6.6 × 105 | abhydrolase domain containing 5, lysophosphatidic acid acyltransferase | ABHD5 |
| ENSSSCG00000015140 | 293 | 1.38 | 8.7 × 109 | heat shock protein family A (Hsp70) member 8 | HSPA8 |
| ENSSSCG00000030095 | 78 | 1.49 | 2.4 × 104 | zinc finger and BTB domain containing 16 | ZBTB16 |
| ENSSSCG00000020754 | 32 | 1.51 | 6.5 × 105 | RNA polymerase I subunit G | CD3EAP |
| ENSSSCG00000005601 | 1136 | 1.54 | 1.8 × 1013 | heat shock protein family A (Hsp70) member 5 | HSPA5 |
| ENSSSCG00000022126 | 97 | 1.73 | 4.0 × 1013 | epidermal growth factor receptor | EGFR |
| ENSSSCG00000010686 | 369 | 1.79 | 8.3 × 106 | BAG cochaperone 3 | BAG3 |
| ENSSSCG00000035058 | 43 | 1.82 | 6.3 × 106 | phosphotyrosine interaction domain containing 1 | PID1 |
| Rapeseed Oil vs. Coconut Oil | |||||
| ENSSSCG00000023331 | 67 | −1.35 | 0.0000 | ubiquitin like 5 | UBL5 |
| ENSSSCG00000007710 | 75 | −1.14 | 0.0033 | MLX interacting protein lik | MLXIPL |
| ENSSSCG00000027926 | 94 | −1.14 | 0.0062 | formimidoyltransferase cyclodeaminase | FTCD |
| ENSSSCG00000003302 | 193 | −1.01 | 0.0004 | Lysoplasmalogenase | TMEM86B |
| ENSSSCG00000031881 | 48 | −1.01 | 0.0003 | CDC42 small effector 1 | CDC42SE1 |
| ENSSSCG00000035790 | 110 | −0.93 | 0.0048 | BTG anti-proliferation factor 1 | BTG1 |
| ENSSSCG00000006719 | 105 | −0.92 | 0.0013 | hydroxy-delta-5-steroid dehydrogenase, 3 β- and steroid delta-isomerase 1 | HSD3B1 |
| ENSSSCG00000031302 | 46 | −0.91 | 0.0015 | C-terminal binding protein 1 | CTBP1 |
| ENSSSCG00000010627 | 45 | −0.88 | 0.0064 | programmed cell death 4 | PDCD4 |
| ENSSSCG00000013514 | 225 | −0.73 | 0.0064 | LRRCT domain-containing protein | PLIN5 |
| ENSSSCG00000001849 | 163 | −0.65 | 0.0062 | alanyl aminopeptidase, membrane | ANPEP |
| ENSSSCG00000014855 | 870 | −0.62 | 0.0002 | ribosomal protein S3 | RPS3 |
| ENSSSCG00000011000 | 253 | 0.86 | 0.0013 | ||
| ENSSSCG00000022126 | 69 | 0.94 | 0.0007 | epidermal growth factor receptor | EGFR |
| ENSSSCG00000039147 | 108 | 0.96 | 0.0014 | ||
| ENSSSCG00000015140 | 278 | 1.14 | 0.0000 | heat shock protein family A (Hsp70) member 8 | HSPA8 |
| ENSSSCG00000004093 | 290 | 1.14 | 0.0000 | iodotyrosine deiodinase | IYD |
| ENSSSCG00000010686 | 339 | 1.49 | 0.0000 | BAG cochaperone 3 | BAG3 |
| ENSSSCG00000011437 | 132 | 1.50 | 0.0005 | 5′-aminolevulinate synthase 1 | ALAS1 |
| ENSSSCG00000024596 | 69 | 1.62 | 0.0051 | nocturnin | NOCT |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oczkowicz, M.; Szmatoła, T.; Świątkiewicz, M.; Koseniuk, A.; Smołucha, G.; Witarski, W.; Wierzbicka, A. 3′quant mRNA-Seq of Porcine Liver Reveals Alterations in UPR, Acute Phase Response, and Cholesterol and Bile Acid Metabolism in Response to Different Dietary Fats. Genes 2020, 11, 1087. https://doi.org/10.3390/genes11091087
Oczkowicz M, Szmatoła T, Świątkiewicz M, Koseniuk A, Smołucha G, Witarski W, Wierzbicka A. 3′quant mRNA-Seq of Porcine Liver Reveals Alterations in UPR, Acute Phase Response, and Cholesterol and Bile Acid Metabolism in Response to Different Dietary Fats. Genes. 2020; 11(9):1087. https://doi.org/10.3390/genes11091087
Chicago/Turabian StyleOczkowicz, Maria, Tomasz Szmatoła, Małgorzata Świątkiewicz, Anna Koseniuk, Grzegorz Smołucha, Wojciech Witarski, and Alicja Wierzbicka. 2020. "3′quant mRNA-Seq of Porcine Liver Reveals Alterations in UPR, Acute Phase Response, and Cholesterol and Bile Acid Metabolism in Response to Different Dietary Fats" Genes 11, no. 9: 1087. https://doi.org/10.3390/genes11091087