Development and Validation of Sex-Specific Markers in Pelodiscus Sinensis Using Restriction Site-Associated DNA Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and DNA Extraction
2.2. Restriction Site-Associated DNA Sequencing
2.3. RAD-seq Data Analysis
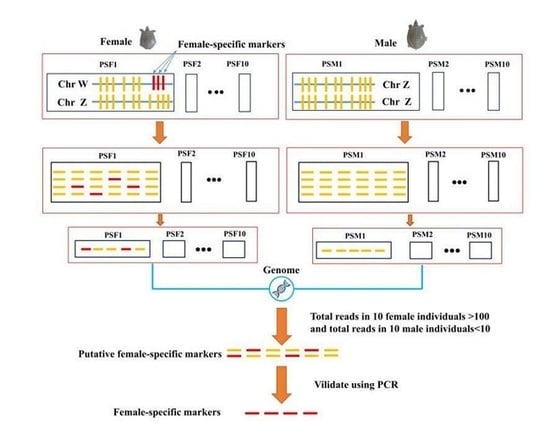
2.4. Sex-Specific Markers Identification
2.5. Validation of Sex-Specific Markers
3. Results
3.1. Restriction Site-Associated DNA Sequencing
3.2. Identification of Sex Specific Regions and Molecular Marker Design
3.3. Identification of Candidate Sex-Difference Regions
3.4. Genome Walking
3.5. Identification of Sex-Specific Markers
3.6. Validation of the Developed Markers
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fritz, U.; Gong, S.; Auer, M.; Kuchling, G.; Schneewei, N.; Jundsdorfer, A.K. The world’s economically most important chelonians represent a diverse species complex (Testudines: Trionychidae: Pelodiscus). Org. Divers. Evol. 2010, 10, 227–242. [Google Scholar] [CrossRef]
- Liang, H.W.; Tong, M.; Cao, L.H.; Li, X.; Zou, G.W. Amino acid and fatty acid composition of three strains of chinese soft-shelled turtle (Pelodiscus sinensis). Pak. J. Zool. 2018, 50, 1061–1069. [Google Scholar] [CrossRef]
- Gong, S.; Vamberger, M.; Auer, M.; Praschag, P.; Fritz, U. Millennium-old farm breeding of chinese softshell turtles (Pelodiscus spp.) results in massive erosion of biodiversity. Sci. Nat. 2018, 105, 34. [Google Scholar] [CrossRef] [PubMed]
- Gui, J.F.; Zhu, Z.Y. Molecular basis and genetic improvement of economically important traits in aquaculture animals. Chin. Sci. Bull. 2012, 57, 1751–1760. [Google Scholar] [CrossRef]
- Zheng, X.H.; Kuang, Y.Y.; Lv, W.H.; Cao, D.C.; Zhang, X.F.; Li, C.; Lu, C.; Sun, X. A consensus linkage map of common carp (Cyprinus carpio L.) to compare the distribution and variation of QTLs associated with growth traits. Sci. China Life Sci. 2013, 56, 351–359. [Google Scholar] [CrossRef]
- Piferrer, F.; Ribas, L.; Díaz, N. Genomic approaches to study genetic and environmental influences on fish sex determination and differentiation. Mar. Biotechnol. 2012, 14, 591–604. [Google Scholar] [CrossRef] [PubMed]
- Sinchan, A.; Soumen, S.; Kumar, B.T.; Parthadeb, G. Identification and validation of a new male sex-specific issr marker in pointed gourd (\r, Trichosanthes dioica\r, roxb.). Sci. World J. 2014, 3, 1–6. [Google Scholar]
- Charlesworth, D.; Mank, J.E. The birds and the bees and the flowers and the trees: Lessons from genetic mapping of sex determination in plants and animals. Genetics 2010, 186, 9–31. [Google Scholar] [CrossRef] [PubMed]
- Fuji, K.; Yoshida, K.; Hattori, K.; Ozaki, A.; Araki, K.; Okauchi, M.; Kubota, S.; Sakamoto, T. Identification of the sex-linked locus in yellowtail, Seriola quinqueradiata. Aquaculture 2010, 308 (Suppl. 1), S51–S55. [Google Scholar] [CrossRef]
- Dan, C.; Mei, J.; Wang, D.; Gui, J.F. Genetic Differentiation and Efficient Sex-specific Marker Development of a Pair of Y- and X-linked Markers in Yellow Catfish. Int. J. Biol. Sci. 2013, 9, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Vale, L.; Dieguez, R.; Sánchez, L.; Martínez, P.; Vinas, A. A sex-associated sequence identified by RAPD screening in gynogenetic individuals of turbot (scophthalmus maximus). Mol. Biol. Rep. 2014, 41, 1501–1509. [Google Scholar] [CrossRef] [PubMed]
- Gamble, T.; Zarkower, D. Identification of sex-specific molecular markers using restriction site-associated DNA sequencing. Mol. Ecol. Resour. 2014, 14, 902–913. [Google Scholar] [CrossRef]
- Pan, Z.J.; Li, X.Y.; Zhou, F.J.; Qiang, X.G.; Gui, J.F. Identification of sex-specific markers reveals male heterogametic sex determination in Pseudobagrus ussuriensis. Mar. Biotechnol. 2015, 17, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.Y.; Xiao, S.J.; Li, W.B.; Wang, Z.Y. Development and validation of sex-specific SNP markers in Larimichthys crocea. J. Fish. China 2018, 42, 1329–1337. [Google Scholar]
- Kawagoshi, T.; Uno, Y.; Matsubara, K.; Matsuda, Y.; Nishida, C. The ZW Micro-Sex Chromosomes of the Chinese Soft-Shelled Turtle (Pelodiscus sinensis, Trionychidae, Testudines) Have the Same Origin as Chicken Chromosome 15. Cytogenet. Genome Res. 2009, 125, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Zhao, B.; Tang, W.Q.; Sun, B.J.; Zeng, Z.G.; Valenzuela, N.; Du, W.G. Temperature-Dependent Sex Determination Ruled Out in the Chinese Soft-Shelled Turtle (Pelodiscus sinensis) via Molecular Cytogenetics and Incubation Experiments across Populations. Sex. Dev. 2015, 9, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Baird, N.A.; Etter, P.D.; Atwood, T.S.; Currey, M.C.; Shiver, A.L.; Lewis, Z.A.; Selker, E.U.; Cresko, W.A.; Johnson, E.A. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 2008, 3, e3376. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Richard, D.B. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Nagahama, Y.; Nakamura, M. Diversity and plasticity of sex determination and differentiation in fishes. Sex. Dev. 2012, 7, 115–125. [Google Scholar] [CrossRef]
- Mei, J.; Gui, J.F. Genetic basis and biotechnological manipulation of sexual dimorphism and sex determination in fish. Sci. China Life Sci. 2015, 58, 124–136. [Google Scholar] [CrossRef]
- Gamble, T.; Coryell, J.; Ezaz, T.; Lynch, J.; Scantlebury, D.P.; Zarkower, D. Restriction site-associated DNA sequencing (RAD-seq) reveals an extraordinary number of transitions among gecko sex-determining systems. Mol. Biol. Evol. 2015, 32, 1296–1309. [Google Scholar] [CrossRef]
- Baxter, S.W.; Davey, J.W.; Johnston, J.S.; Shelton, A.M.; Heckel, D.G.; Jiggins, C.D.; Blaxter, M.L. Linkage mapping and comparative genomics using next-generation rad sequencing of a non-model organism. PLoS ONE 2011, 6, e19315. [Google Scholar] [CrossRef]
- Anderson, J.L.; Marí, R.M.; Braasch, I.; Amores, A.; Hohenlohe, P.; Batzel, P.; Postlethwait, J.H. Multiple sex-associated regions and a putative sex chromosome in zebrafish revealed by rad mapping and population genomics. PLoS ONE 2012, 7, e40701. [Google Scholar] [CrossRef]
- Amores, A.; Catchen, J.; Ferrara, A.; Fontenot, Q.; Postlethwait, J.H. Genome evolution and meiotic maps by massively parallel DNA sequencing: Spotted gar, an outgroup for the teleost genome duplication. Genetics 2011, 188, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Ritland, K. Evolutionary potential in the wild: more than meets the eye. Mol. Ecol. 2011, 20, 3494–3495. [Google Scholar] [CrossRef] [PubMed]
- Fowler, B.L.S.; Buonaccorsi, V.P. Genomic characterization of sex-identification markers in Sebastes carnatus and S. chrysomelas rockfishes. Mol. Ecol. 2016, 25, 2165–2175. [Google Scholar] [CrossRef] [PubMed]
- Alkan, C.; Coe, B.P.; Eichler, E.E. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 2011, 12, 363–376. [Google Scholar] [CrossRef] [PubMed]
- Baker, M. Structural variation: The genome’s hidden architecture. Nat. Methods 2012, 9, 133–137. [Google Scholar] [CrossRef]
- Veitia, R.A.; Bottani, S.; Birchler, J.A. Gene dosage effects: Nonlinearities, genetic interactions, and dosage compensation. Trends Genet. Tig. 2013, 29, 385–393. [Google Scholar] [CrossRef] [PubMed]






| Locus | Markers | Sequence (5′−3′) | Purpose |
|---|---|---|---|
| ps4085 | ps4085F | GTTTGAAGTGCTGCTGGGAAG | Identification of the candidate female-specific marker region |
| ps4085R | TTCCCCGTATAAAGCCAGGG | ||
| ps3137 | ps3137F | CACGGGGTGAAGCACTCATT | |
| ps3137R | TGTCTCACCTCCTTTGCCTC | ||
| ps2962 | ps2926F | AGAATTCATGCGGTTACTCAACT | |
| ps2926R | GAGGCAGCATTTGACGGGTA | ||
| ps5510 | ps5510F | GGGAAATAACGGGACCCCAG | |
| ps5510R | CCACTGGTATCTGGCCACTC | ||
| ps5388 | ps5388F | GGAATTCTGCGGGTAGGAGG | |
| ps5388R | CCTAACTCCTGGGGGAACCT | ||
| ps7307 | ps7307F | GCCAGGACACTGGCATTAAC | |
| ps7307R | GAGGTCCTGGACACCTCTGA | ||
| ps7885 | ps7885F | CTCCATAGGGGAGCACAAGC | |
| ps7885R | TGCAGGTGCAGAACCACTTA | ||
| ps7837 | ps7837F | GGTCTTAATGCACCCCGTTG | |
| ps7837R | ACATTTCCCACCCCTTGGTC | ||
| ps5946 | ps5946F | TTAGGTGCCTAGTGCCAAGC | |
| ps5946R | CCCGTCCCGAAGACTGTATG | ||
| ps6704 | ps6704F | CCCAACTGCATTCCCGTTCT | |
| ps6704R | CCCTGGACGTGGAAAACTCA | ||
| ps4085s | ps4085sF | GCTGCTGGGAAGACTTAGAGTTG | Identification of sex-specific marker |
| ps4085sR | CTTGAATGTGACTAGGAAGGCTTC | ||
| ps3137s1 | ps3137s1F | TGACTGGCACTGGAGAAGGA | |
| ps3137s1R | GCATCAACCACAGCCCTACA | ||
| ps3137s2 | ps3137s2F | TCCAGGGCAGAAAGACACTC | |
| ps3137s2R | ATGTCCCGGTGGATGGC |
| Sample | Raw Data bp | Raw Reads Num. | Q20% | Q30% | GC% | HQ Reads Num (%) |
|---|---|---|---|---|---|---|
| PSM1 | 2,957,269,944 | 21,351,164 | 96.46 | 92.10 | 43.67 | 20,687,143 (96.89%) |
| PSM2 | 2,411,667,340 | 16,504,674 | 96.89 | 92.68 | 43.32 | 16,120,788 (97.67%) |
| PSM3 | 2,595,681,148 | 17,558,578 | 96.07 | 91.17 | 43.98 | 16,952,904 (96.55%) |
| PSM4 | 2,226,436,872 | 15,037,496 | 95.88 | 90.87 | 44.43 | 14,450,386 (96.10%) |
| PSM5 | 2,387,165,348 | 16,221,390 | 96.14 | 91.32 | 44.47 | 15,691,924 (96.74%) |
| PSM6 | 2,054,437,868 | 13,875,546 | 95.94 | 90.87 | 43.82 | 13,385,482 (96.47%) |
| PSM7 | 2,481,263,740 | 16,868,546 | 96.22 | 91.44 | 44.07 | 16,346,936 (96.91%) |
| PSM8 | 2,001,024,316 | 13,705,646 | 96.55 | 92.02 | 44.33 | 13,397,652 (97.75%) |
| PSM9 | 1,987,020,528 | 13,411,628 | 95.86 | 90.84 | 44.43 | 12,877,632 (96.02%) |
| PSM10 | 2,307,425,680 | 15,574,296 | 95.85 | 90.72 | 44.10 | 14,991,562 (96.26%) |
| PSF1 | 2,100,080,936 | 14,384,116 | 96.62 | 92.28 | 44.17 | 14,034,716 (97.57%) |
| PSF2 | 2,467,693,044 | 16,759,374 | 96.44 | 91.92 | 43.89 | 16,275,792 (97.11%) |
| PSF3 | 2,213,291,672 | 15,159,532 | 96.80 | 92.60 | 43.95 | 14,830,036 (97.83%) |
| PSF4 | 2,201,335,428 | 14,891,202 | 96.28 | 91.59 | 43.88 | 14,393,778 (96.66%) |
| PSF5 | 2,714,080,420 | 18,338,706 | 95.96 | 91.17 | 44.06 | 17,563,492 (95.77%) |
| PSF6 | 3,250,793,660 | 22,265,710 | 96.77 | 92.52 | 43.89 | 21,795,770 (97.89%) |
| PSF7 | 2,429,666,592 | 16,641,552 | 96.72 | 92.46 | 44.02 | 16,261,182 (97.71%) |
| PSF8 | 3,582,429,740 | 24,537,190 | 96.13 | 91.90 | 43.89 | 23,398,702 (95.36%) |
| PSF9 | 2,650,642,556 | 18,155,086 | 96.86 | 92.70 | 43.83 | 17,784,484 (97.96%) |
| PSF10 | 2,673,816,280 | 18,136,448 | 96.45 | 91.86 | 43.77 | 17,644,914 (97.29%) |
| Sample | All Reads | Single Mapped Reads | Paired Mapped Reads | Unmapped Reads | Alignment Ratio (%) |
|---|---|---|---|---|---|
| PSM1 | 20,687,143 | 154,583 | 12,937,651 | 422,017 | 97.96 |
| PSM2 | 16,120,788 | 52,145 | 7,866,467 | 335,709 | 97.92 |
| PSM3 | 16,952,904 | 124,324 | 8,213,599 | 401,382 | 97.63 |
| PSM4 | 14,450,386 | 108,720 | 7,007,918 | 325,830 | 97.75 |
| PSM5 | 15,691,924 | 121,822 | 7,592,756 | 384,590 | 97.55 |
| PSM6 | 13,385,482 | 98,822 | 6,511,777 | 263,106 | 98.03 |
| PSM7 | 16,346,936 | 116,368 | 7,961,448 | 307,672 | 98.12 |
| PSM8 | 13,397,652 | 97,082 | 6,530,704 | 239,162 | 98.21 |
| PSM9 | 12,877,632 | 99,272 | 6,224,305 | 329,750 | 97.44 |
| PSM10 | 14,991,562 | 113,334 | 7,283,306 | 311,616 | 97.92 |
| PSF1 | 14,034,716 | 910,48 | 6,857,225 | 229,218 | 98.37 |
| PSF2 | 16,275,792 | 106,920 | 7,934,999 | 298,874 | 98.16 |
| PSF3 | 14,830,036 | 92,616 | 7,229,123 | 279,174 | 98.12 |
| PSF4 | 14,393,778 | 92,498 | 7,026,635 | 248,010 | 98.28 |
| PSF5 | 17,563,492 | 121,625 | 8,549,820 | 342,227 | 98.05 |
| PSF6 | 21,795,770 | 142,215 | 10,640,276 | 373,003 | 98.29 |
| PSF7 | 16,261,182 | 107,657 | 7,921,154 | 311,217 | 98.09 |
| PSF8 | 23,398,702 | 138,420 | 11,436,319 | 387,644 | 98.34 |
| PSF9 | 17,784,484 | 109,638 | 8,697,230 | 280,386 | 98.42 |
| PSF10 | 17,644,914 | 112,739 | 8,617,538 | 297,099 | 98.32 |
| Name | Size | Frequency (%) | |||
|---|---|---|---|---|---|
| Hanshou Population | Xiantao Population | ||||
| Female | Male | Female | Male | ||
| ps4085 | 1434 bp | 48/48 (100) | 48/48 (100) | 100/100 (100) | 100/100 (100) |
| ps3137s1 | 1313 bp | 48/48 (100) | 48/48 (100) | 100/100 (100) | 100/100 (100) |
| ps3137s2 | 864 bp | 48/48 (100) | 48/48 (100) | 100/100 (100) | 100/100 (100) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, H.; Wang, L.; Sha, H.; Zou, G. Development and Validation of Sex-Specific Markers in Pelodiscus Sinensis Using Restriction Site-Associated DNA Sequencing. Genes 2019, 10, 302. https://doi.org/10.3390/genes10040302
Liang H, Wang L, Sha H, Zou G. Development and Validation of Sex-Specific Markers in Pelodiscus Sinensis Using Restriction Site-Associated DNA Sequencing. Genes. 2019; 10(4):302. https://doi.org/10.3390/genes10040302
Chicago/Turabian StyleLiang, Hongwei, Lihua Wang, Hang Sha, and Guiwei Zou. 2019. "Development and Validation of Sex-Specific Markers in Pelodiscus Sinensis Using Restriction Site-Associated DNA Sequencing" Genes 10, no. 4: 302. https://doi.org/10.3390/genes10040302