In Vitro-Generated Hypertrophic-Like Adipocytes Displaying PPARG Isoforms Unbalance Recapitulate Adipocyte Dysfunctions In Vivo
Abstract
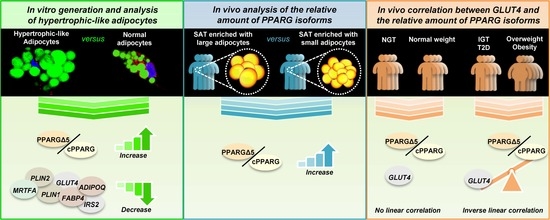
:1. Introduction
2. Materials and Methods
2.1. Human Samples
2.2. Cell Lines and Cultures
2.3. In Vitro Differentiation of Mature and Hypertrophic-Like Adipocytes
2.4. Immunofluorescence Microscopy
2.5. Cell Count and Oil Red O Staining for Quantifying Adipocyte Differentiation
2.6. Analysis of Adipocyte Size and Lipid Droplets
2.7. Transmission Electron Microscopy
2.8. RNA Extraction, RT-PCR and qPCR
2.9. Western Blot
2.10. Flow Cytometry Analysis
2.11. ELISA
2.12. Quantification and Statistical Analysis
3. Results and Discussion
3.1. Unbalance of PPARG Isoforms in Patients with Hypertrophic Obesity
3.2. From Human Mesenchymal Stem Cells to Mature Adipocytes
3.3. From Mature to Hypertrophic Adipocytes
3.4. Stepwise Expression from hMSCs to Hypertrophic-Like Adipocytes
3.5. Hypertrophic-Like Adipocytes Display Morphological Features Resembling Adipose Tissue Hypertrophy
3.6. GLUT4 Negatively Correlates with PPARGΔ5/cPPARG Ratio Only in Pathologic Conditions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Danforth, E. Failure of adipocyte differentiation causes type II diabetes mellitus? Nat. Genet. 2000, 26, 13. [Google Scholar] [CrossRef] [PubMed]
- Shulman, G.I. Cellular mechanisms of insulin resistance. J. Clin. Investig. 2000, 106, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Savage, D.B.; Petersen, K.F.; Shulman, G.I. Disordered lipid metabolism and the pathogenesis of insulin resistance. Physiol. Rev. 2007, 87, 507–520. [Google Scholar] [CrossRef] [PubMed]
- Savage, D.B. Mouse models of inherited lipodystrophy. Dis. Model. Mech. 2009, 2, 554–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gustafson, B.; Hammarstedt, A.; Hedjazifar, S.; Smith, U. Restricted Adipogenesis in Hypertrophic Obesity. Diabetes 2013, 62, 2997–3004. [Google Scholar] [CrossRef] [Green Version]
- Muir, L.; Neeley, C.K.; Meyer, K.A.; Baker, N.A.; Brosius, A.M.; Washabaugh, A.R.; Varban, O.A.; Finks, J.F.; Zamarron, B.F.; Flesher, C.G.; et al. Adipose tissue fibrosis, hypertrophy, and hyperplasia: Correlations with diabetes in human obesity. Obesity 2016, 24, 597–605. [Google Scholar] [CrossRef]
- Klöting, N.; Blüher, M. Adipocyte dysfunction, inflammation and metabolic syndrome. Rev. Endocr. Metab. Disord. 2014, 15, 277–287. [Google Scholar] [CrossRef]
- Savage, D.B. PPAR[gamma] as a metabolic regulator: Insights from genomics and pharmacology. Expert Rev. Mol. Med. 2005, 7. [Google Scholar] [CrossRef]
- Sadowski, H.B.; Wheeler, T.T.; Young, D.A. Gene expression during 3T3-L1 adipocyte differentiation. Characterization of initial responses to the inducing agents and changes during commitment to differentiation. J. Boil. Chem. 1992, 267, 4722–4731. [Google Scholar]
- Kim, J.I.; Huh, J.Y.; Sohn, J.H.; Choe, S.S.; Lee, Y.S.; Lim, C.Y.; Jo, A.; Park, S.B.; Han, W.; Kim, J.B. Lipid-Overloaded Enlarged Adipocytes Provoke Insulin Resistance Independent of Inflammation. Mol. Cell. Boil. 2015, 35, 1686–1699. [Google Scholar] [CrossRef] [Green Version]
- Cawthorn, W.P.; Scheller, E.L.; MacDougald, O. Adipose tissue stem cells meet preadipocyte commitment: Going back to the future[S]. J. Lipid Res. 2011, 53, 227–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Ordemann, J.; Müller, J.M.; Dubiel, W. The COP9 signalosome, cullin 3 and Keap1 super complex regulates CHOP stability and adipogenesis. Biol. Open 2012, 1, 705–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunnell, B.A.; Flaat, M.; Gagliardi, C.; Patel, B.; Ripoll, C. Adipose-derived stem cells: Isolation, expansion and differentiation☆. Methods 2008, 45, 115–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Ojeda, F.J.; Iris-Rupérez, A.; Gómez-Llorente, C.; Gil, A.; Aguilera, C.M. Cell Models and Their Application for Studying Adipogenic Differentiation in Relation to Obesity: A Review. Int. J. Mol. Sci. 2016, 17, 1040. [Google Scholar] [CrossRef] [Green Version]
- Mohamed-Ahmed, S.; Fristad, I.; Lie, S.A.; Suliman, S.; Mustafa, K.; Vindenes, H.; Idris, S.B. Adipose-derived and bone marrow mesenchymal stem cells: A donor-matched comparison. Stem Cell Res. Ther. 2018, 9, 168. [Google Scholar] [CrossRef]
- Aprile, M.; Cataldi, S.; Ambrosio, M.R.; D’Esposito, V.; Lim, K.; Dietrich, A.; Blüher, M.; Savage, D.B.; Formisano, P.; Ciccodicola, A.; et al. PPARγΔ5, a Naturally Occurring Dominant-Negative Splice Isoform, Impairs PPARγ Function and Adipocyte Differentiation. Cell Rep. 2018, 25, 1577–1592. [Google Scholar] [CrossRef] [Green Version]
- Stenkula, K.G.; Erlanson-Albertsson, C. Adipose cell size: Importance in health and disease. Am. J. Physiol. Integr. Comp. Physiol. 2018, 315, R284–R295. [Google Scholar] [CrossRef] [Green Version]
- Costa, V.; Gallo, M.A.; Letizia, F.; Aprile, M.; Casamassimi, A.; Ciccodicola, A. PPARG: Gene Expression Regulation and Next-Generation Sequencing for Unsolved Issues. PPAR Res. 2010, 2010, 1–17. [Google Scholar] [CrossRef]
- Aprile, M.; Ambrosio, M.R.; D’Esposito, V.; Beguinot, F.; Formisano, P.; Costa, V.; Ciccodicola, A. PPARG in human adipogenesis: Differential contribution of canonical transcripts and dominant negative isoforms. PPAR Res. 2014, 2014, 18–20. [Google Scholar] [CrossRef] [Green Version]
- Keller, M.; Hopp, L.; Liu, X.; Wohland, T.; Rohde, K.; Cancello, R.; Klös, M.; Bacos, K.; Kern, M.; Eichelmann, F.; et al. Genome-wide DNA promoter methylation and transcriptome analysis in human adipose tissue unravels novel candidate genes for obesity. Mol. Metab. 2016, 6, 86–100. [Google Scholar] [CrossRef]
- Guiu-Jurado, E.; Unthan, M.; Böhler, N.; Kern, M.; Landgraf, K.; Dietrich, A.; Schleinitz, R.; Ruschke, K.; Klöting, N.; Faßhauer, M.; et al. Bone morphogenetic protein 2 (BMP2 ) may contribute to partition of energy storage into visceral and subcutaneous fat depots. Obesity 2016, 24, 2092–2100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Isakson, P.; Hammarstedt, A.; Gustafson, B.; Smith, U. Impaired Preadipocyte Differentiation in Human Abdominal Obesity. Diabetes 2009, 58, 1550–1557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armoni, M.; Harel, C.; Karnieli, E. Transcriptional regulation of the GLUT4 gene: From PPAR-γ and FOXO1 to FFA and inflammation. Trends Endocrinol. Metab. 2007, 18, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Janderová, L.; McNeil, M.; Murrell, A.; Mynatt, R.L.; Smith, S.R.; Rossmeislová, L. Human Mesenchymal Stem Cells as an in Vitro Model for Human Adipogenesis. Obes. Res. 2003, 11, 65–74. [Google Scholar] [CrossRef] [PubMed]
- McBeath, R.; Pirone, D.M.; Nelson, C.M.; Bhadriraju, K.; Chen, C.S. Cell Shape, Cytoskeletal Tension, and RhoA Regulate Stem Cell Lineage Commitment. Dev. Cell 2004, 6, 483–495. [Google Scholar] [CrossRef] [Green Version]
- Modica, S.; Wolfrum, C. The dual role of BMP4 in adipogenesis and metabolism. Adipocyte 2017, 6, 141–146. [Google Scholar] [CrossRef] [Green Version]
- Walther, T.C.; Chung, J.; Farese, R.V. Lipid Droplet Biogenesis. Annu. Rev. Cell Dev. Boil. 2017, 33, 491–510. [Google Scholar] [CrossRef] [Green Version]
- Heid, H.; Rickelt, S.; Zimbelmann, R.; Winter, S.; Schumacher, H.; Dörflinger, Y.; Kuhn, C.; Franke, W. On the Formation of Lipid Droplets in Human Adipocytes: The Organization of the Perilipin–Vimentin Cortex. PLoS ONE 2014, 9, e90386. [Google Scholar] [CrossRef] [Green Version]
- Khatau, S.B.; Hale, C.; Stewart-Hutchinson, P.J.; Patel, M.S.; Stewart, C.L.; Searson, P.; Hodzic, D.; Wirtz, D. A perinuclear actin cap regulates nuclear shape. Proc. Natl. Acad. Sci. USA 2009, 106, 19017–19022. [Google Scholar] [CrossRef] [Green Version]
- Spiegelman, B.M.; Hu, E.; Kim, J.B.; Brun, R. PPAR gamma and the control of adipogenesis. Biochimie 1997, 79, 111–112. [Google Scholar]
- Costa, V.; Ciccodicola, A. Is PPARG the key gene in diabetic retinopathy? Br. J. Pharmacol. 2012, 165, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Tontonoz, P.; Spiegelman, B.M. Fat and Beyond: The Diverse Biology of PPARγ. Annu. Rev. Biochem. 2008, 77, 289–312. [Google Scholar] [CrossRef]
- Prokesch, A.; Smorlesi, A.; Perugini, J.; Manieri, M.; Ciarmela, P.; Mondini, E.; Trajanoski, Z.; Kristiansen, K.; Giordano, A.; Bogner-Strauss, J.G.; et al. Molecular aspects of adipoepithelial transdifferentiation in mouse mammary gland. STEM CELLS 2014, 32, 2756–2766. [Google Scholar] [CrossRef] [PubMed]
- Wolins, N.E.; Quaynor, B.K.; Skinner, J.R.; Schoenfish, M.J.; Tzekov, A.; Bickel, P.E. S3-12, Adipophilin, and TIP47 Package Lipid in Adipocytes. J. Boil. Chem. 2005, 280, 19146–19155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalen, K.T.; Schoonjans, K.; Ulven, S.M.; Weedon-Fekjaer, M.S.; Bentzen, T.G.; Koutnikova, H.; Auwerx, J.; Nebb, H.I. Adipose tissue expression of the lipid droplet-associating proteins S3-12 and perilipin is controlled by peroxisome proliferator-activated receptor-gamma. Diabetes 2004, 53. [Google Scholar] [CrossRef] [Green Version]
- Arimura, N.; Horiba, T.; Imagawa, M.; Shimizu, M.; Sato, R. The Peroxisome Proliferator-activated Receptor γ Regulates Expression of the Perilipin Gene in Adipocytes. J. Boil. Chem. 2004, 279, 10070–10076. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, Y.; Shinoda, A.; Kamada, H.; Shimizu, M.; Inoue, J.; Sato, R. Perilipin2 plays a positive role in adipocytes during lipolysis by escaping proteasomal degradation. Sci. Rep. 2016, 6, 20975. [Google Scholar] [CrossRef] [Green Version]
- Itabe, H.; Yamaguchi, T.; Nimura, S.; Sasabe, N. Perilipins: A diversity of intracellular lipid droplet proteins. Lipids Heal. Dis. 2017, 16, 83. [Google Scholar] [CrossRef] [Green Version]
- Kanzaki, M.; Pessin, J.E. Insulin-stimulated GLUT4 Translocation in Adipocytes Is Dependent upon Cortical Actin Remodeling. J. Boil. Chem. 2001, 276, 42436–42444. [Google Scholar] [CrossRef] [Green Version]
- Nobusue, H.; Onishi, N.; Shimizu, T.; Sugihara, E.; Oki, Y.; Sumikawa, Y.; Chiyoda, T.; Akashi, K.; Saya, H.; Kano, K. Regulation of MKL1 via actin cytoskeleton dynamics drives adipocyte differentiation. Nat. Commun. 2014, 5, 3368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gustafson, B.; Gogg, S.; Hedjazifar, S.; Jenndahl, L.; Hammarstedt, A.; Smith, U. Inflammation and impaired adipogenesis in hypertrophic obesity in man. Am. J. Physiol. Metab. 2009, 297, E999–E1003. [Google Scholar] [CrossRef] [Green Version]
- Gustafson, B.; Hedjazifar, S.; Gogg, S.; Hammarstedt, A.; Smith, U. Insulin resistance and impaired adipogenesis. Trends Endocrinol. Metab. 2015, 26, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Hajer, G.R.; Van Haeften, T.W.; Visseren, F.L. Adipose tissue dysfunction in obesity, diabetes, and vascular diseases. Eur. Hear. J. 2008, 29, 2959–2971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugii, S.; Olson, P.; Sears, R.D.; Saberi, M.; Atkins, A.R.; Barish, G.D.; Hong, S.-H.; Castro, G.L.; Yin, Y.-Q.; Nelson, M.C.; et al. PPARgamma activation in adipocytes is sufficient for systemic insulin sensitization. Proc. Natl. Acad. Sci. USA 2009, 106, 22504–22509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, U.; Gogg, S.; Johansson, A.; Olausson, T.; Rotter, V.; Svalstedt, B. Thiazolidinediones (PPARγ agonists) but not PPAR α agonists increase IRS-2 gene expression in 3T3-L1 and human adipocytes 1. FASEB J. 2001, 15, 215–220. [Google Scholar] [CrossRef]
- Lieber, J.G.; Evans, R.M. Disruption of the vimentin intermediate filament system during adipose conversion of 3T3-L1 cells inhibits lipid droplet accumulation. J. Cell Sci. 1996, 109, 109. [Google Scholar]
- McGookey, D.J.; Anderson, R.G. Morphological characterization of the cholesteryl ester cycle in cultured mouse macrophage foam cells. J. Cell Boil. 1983, 97, 1156–1168. [Google Scholar] [CrossRef] [Green Version]
- Murphy, D.J.; Vance, J. Mechanisms of lipid-body formation. Trends Biochem. Sci. 1999, 24, 109–115. [Google Scholar] [CrossRef]
- Van Meer, G. Caveolin, Cholesterol, and Lipid Droplets? J. Cell Boil. 2001, 152, F29–F34. [Google Scholar] [CrossRef] [Green Version]
- Thiam, A.R.; Farese, R.V., Jr.; Walther, T.C. The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Boil. 2013, 14, 775–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fei, W.; Shui, G.; Zhang, Y.; Krahmer, N.; Ferguson, C.; Kapterian, T.S.; Lin, R.C.Y.; Dawes, I.W.; Brown, A.J.; Li, P.; et al. A Role for Phosphatidic Acid in the Formation of “Supersized” Lipid Droplets. PLoS Genet. 2011, 7, e1002201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skurk, T.; Alberti-Huber, C.; Herder, C.; Hauner, H. Relationship between Adipocyte Size and Adipokine Expression and Secretion. J. Clin. Endocrinol. Metab. 2007, 92, 1023–1033. [Google Scholar] [CrossRef] [PubMed]
- Almuraikhy, S.; Kafienah, W.; Bashah, M.; Diboun, I.; Jaganjac, M.; Al-Khelaifi, F.; Abdesselem, H.; Mazloum, N.A.; Alsayrafi, M.; Mohamed-Ali, V.; et al. Interleukin-6 induces impairment in human subcutaneous adipogenesis in obesity-associated insulin resistance. Diabetologia 2016, 59, 2406–2416. [Google Scholar] [CrossRef] [Green Version]
- Rotter, V.; Nagaev, I.; Smith, U. Interleukin-6 (IL-6) Induces Insulin Resistance in 3T3-L1 Adipocytes and Is, Like IL-8 and Tumor Necrosis Factor-α, Overexpressed in Human Fat Cells from Insulin-resistant Subjects. J. Boil. Chem. 2003, 278, 45777–45784. [Google Scholar] [CrossRef] [Green Version]
- Bastard, J.-P.; Maachi, M.; van Nhieu, J.T.; Jardel, C.; Bruckert, E.; Grimaldi, A.; Robert, J.-J.; Capeau, J.; Hainque, B. Adipose Tissue IL-6 Content Correlates with Resistance to Insulin Activation of Glucose Uptake both in Vivo and in Vitro. J. Clin. Endocrinol. Metab. 2002, 87, 2084–2089. [Google Scholar] [CrossRef]
- Kern, P.A.; Ranganathan, S.; Li, C.; Wood, L.; Ranganathan, G. Adipose tissue tumor necrosis factor and interleukin-6 expression in human obesity and insulin resistance. Am. J. Physiol. Metab. 2001, 280, E745–E751. [Google Scholar] [CrossRef]
- Sadur, U.N.; Yost, T.J.; Eckel, R.H. Insulin Responsiveness of Adipose Tissue Lipoprotein Lipase Is Delayed but Preserved in Obesity*. J. Clin. Endocrinol. Metab. 1984, 59, 1176–1182. [Google Scholar] [CrossRef]
- Serra, M.C.; Ryan, A.S.; Sorkin, J.D.; Favor, K.H.; Goldberg, A.P.; Favors, K.H. High adipose LPL activity and adipocyte hypertrophy reduce visceral fat and metabolic risk in obese, older women. Obesity 2015, 23, 602–607. [Google Scholar] [CrossRef] [Green Version]
- Farnier, C.; Krief, S.; Blache, M.; Diot-Dupuy, F.; Mory, G.; Ferre, P.; Bazin, R. Adipocyte functions are modulated by cell size change: Potential involvement of an integrin/ERK signalling pathway. Int. J. Obes. 2003, 27, 1178–1186. [Google Scholar] [CrossRef] [Green Version]
- McTernan, P.G.; Harte, A.L.; Anderson, L.A.; Green, A.; Smith, S.A.; Holder, J.C.; Barnett, A.H.; Eggo, M.C.; Kumar, S. Insulin and rosiglitazone regulation of lipolysis and lipogenesis in human adipose tissue in vitro. Diabetes 2002, 51, 1493–1498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sohn, J.H.; Lee, Y.K.; Han, J.S.; Jeon, Y.G.; Kim, J.I.; Choe, S.S.; Kim, S.J.; Yoo, H.J.; Kim, J.B. Perilipin 1 (Plin1) deficiency promotes inflammatory responses in lean adipose tissue through lipid dysregulation. J. Boil. Chem. 2018, 293, 13974–13988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czech, M.P. Insulin action and resistance in obesity and type 2 diabetes. Nat. Med. 2017, 23, 804–814. [Google Scholar] [CrossRef] [PubMed]







| Women | Men | |
|---|---|---|
| n | 51 | 35 |
| Age (years) | 53.4 ± 16.6 | 58.4 ± 16.3 |
| Body weight (kg) | 99.5 ± 34.4 | 109 ± 44.9 |
| BMI (kg/m²) | 36.5 ± 12 | 34.1 ± 12.3 |
| Body fat (%) | 35 ± 11.7 | 30.7 ± 9.5 |
| Visceral fat area (cm²) | 165.5 ± 119.6 | 172.9 ± 134 |
| Subcutaneous fat area (cm²) | 471 ± 492.5 | 418.5 ± 333.3 |
| Waist circumference (cm) | 114 ± 33.1 | 120.3 ± 24 |
| FPG (mmol/L) | 5.9 ± 1.2 | 5.7 ± 0.9 |
| FPI (pmol/L) | 127.5 ± 133.1 | 81.4 ± 89.8 |
| HbA1c (%) | 5.9 ± 0.8 | 5.9 ± 0.62 |
| Clamp GIR (µmol/kg/min) | 79 ± 35 | 75.2 ± 32 |
| Cholesterol (mmol/L) | 5.1 ± 0.75 | 4.9 ± 1.02 |
| HDL-Cholesterol (mmol/L) | 1.2 ± 0.3 | 1.1 ± 0.3 |
| LDL-Cholesterol (mmol/L) | 3.6 ± 1.2 | 3.5 ± 1.2 |
| Triglycerides (mmol/L) | 1.42 ± 0.36 | 1.9 ± 1.6 |
| Free fatty acids (mmol/L) | 0.44 ± 0.38 | 0.47 ± 0.4 |
| hsCRP (mg/L) | 12.3 ± 14.8 | 11.5 ± 14.1 |
| IL-6 (pg/mL) | 4.2 ± 4.1 | 3.3 ± 4.4 |
| ALAT (µkat/L) | 0.8 ± 1.1 | 0.67 ± 0.7 |
| ASAT (µkat/L) | 0.7 ± 0.85 | 0.63 ± 0.5 |
| GGT (µkat/L) | 1.9 ± 3.5 | 1.5 ± 2.5 |
| Adiponectin (µg/mL) | 9.1 ± 6.6 | 5 ± 3** |
| Leptin (pg/mL) | 40 ± 20 | 18.5 ± 11.5** |
| Mean subcutaneous adipocyte diameter (µm) | 110.6 ± 11.6 | 107.6 ± 9.3 |
| Mean visceral adipocyte diameter (µm) | 100 ± 7.4 | 97.6 ± 5.7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aprile, M.; Cataldi, S.; Perfetto, C.; Ambrosio, M.R.; Italiani, P.; Tatè, R.; Blüher, M.; Ciccodicola, A.; Costa, V. In Vitro-Generated Hypertrophic-Like Adipocytes Displaying PPARG Isoforms Unbalance Recapitulate Adipocyte Dysfunctions In Vivo. Cells 2020, 9, 1284. https://doi.org/10.3390/cells9051284
Aprile M, Cataldi S, Perfetto C, Ambrosio MR, Italiani P, Tatè R, Blüher M, Ciccodicola A, Costa V. In Vitro-Generated Hypertrophic-Like Adipocytes Displaying PPARG Isoforms Unbalance Recapitulate Adipocyte Dysfunctions In Vivo. Cells. 2020; 9(5):1284. https://doi.org/10.3390/cells9051284
Chicago/Turabian StyleAprile, Marianna, Simona Cataldi, Caterina Perfetto, Maria Rosaria Ambrosio, Paola Italiani, Rosarita Tatè, Matthias Blüher, Alfredo Ciccodicola, and Valerio Costa. 2020. "In Vitro-Generated Hypertrophic-Like Adipocytes Displaying PPARG Isoforms Unbalance Recapitulate Adipocyte Dysfunctions In Vivo" Cells 9, no. 5: 1284. https://doi.org/10.3390/cells9051284