Insertion Defects of Mitochondrially Encoded Proteins Burden the Mitochondrial Quality Control System
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Growth Media
2.2. Paromomycin Disk Diffusion Assay
2.3. Isolation of Mitochondria
2.4. Labeling of Mitochondrial Translation Products in Organello
3. Results
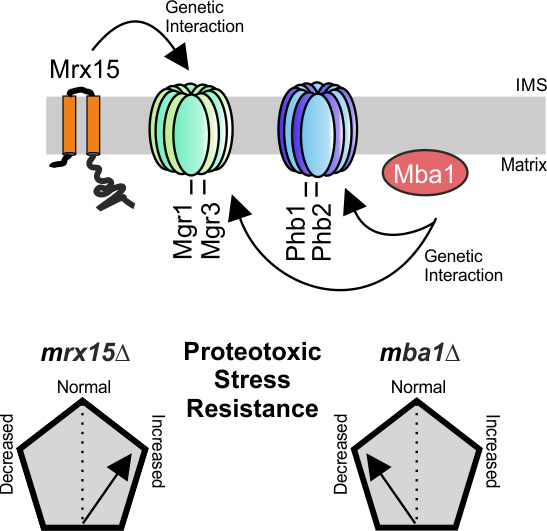
3.1. MRX15 Deletion Causes a Proteotoxic Resistant Phenotype
3.2. MRX15 and MBA1 Functionally Interact with Regulators of the i-AAA Protease
3.3. Mgr1 and Mgr3 are Required during Proteotoxic Stress
3.4. MRX15 is Specifically Linked to the i-AAA Protease
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Burger, G.; Gray, M.W.; Lang, B.F. Mitochondrial genomes: Anything goes. Trends Genet. 2003, 19, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Bestwick, M.L.; Shadel, G.S. Accessorizing the human mitochondrial transcription machinery. Trends Biochem. Sci. 2013, 38, 283–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ott, M.; Amunts, A.; Brown, A. Organization and regulation of mitochondrial protein synthesis. Annu. Rev. Biochem. 2016, 85, 77–101. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, K.A.; Kaniak-Golik, A.; Golik, P. Maintenance and expression of the S. cerevisiae mitochondrial genome—From genetics to evolution and systems biology. Biochim. Biophys. Acta 2010, 1797, 1086–1098. [Google Scholar] [CrossRef] [PubMed]
- Voos, W. Chaperone—Protease networks in mitochondrial protein homeostasis. Biochim. Biophys. Acta 2013, 1833, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, F.; Tatsuta, T.; Langer, T. Mitochondrial AAA proteases—Towards a molecular understanding of membrane-bound proteolytic machines. Biochim. Biophys. Acta Mol. Cell Res. 2012, 1823, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Weber, E.R.; Hanekamp, T.; Thorsness, P.E. Biochemical and functional analysis of the YME1 gene product, an ATP and zinc-dependent mitochondrial protease from S. cerevisiae. Mol. Biol. Cell 1996, 7, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Arlt, H.; Tauer, R.; Feldmann, H.; Neupert, W.; Langer, T. The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 1996, 85, 875–885. [Google Scholar] [CrossRef]
- Puchades, C.; Rampello, A.J.; Shin, M.; Giuliano, C.J.; Wiseman, R.L.; Glynn, S.E.; Lander, G.C. Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science 2017, 358, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Leonhard, K.; Herrmann, J.M.; Stuart, R.A.; Mannhaupt, G.; Neupert, W.; Langer, T. AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J. 1996, 15, 4218–4229. [Google Scholar] [CrossRef] [PubMed]
- Dunn, C.D.; Tamura, Y.; Sesaki, H.; Jensen, R.E. Mgr3p and Mgr1p are adaptors for the mitochondrial i-AAA protease complex. Mol. Biol. Cell 2008, 19, 5387–5397. [Google Scholar] [CrossRef] [PubMed]
- Dunn, C.D.; Lee, M.S.; Spencer, F.A.; Jensen, R.E. A genomewide screen for petite-negative yeast strains yields a new subunit of the i-AAA protease complex. Mol. Biol. Cell 2006, 17, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Steglich, G.; Neupert, W.; Langer, T. Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol. Cell. Biol. 1999, 19, 3435–3442. [Google Scholar] [CrossRef] [PubMed]
- Merkwirth, C.; Langer, T. Prohibitin function within mitochondria: Essential roles for cell proliferation and cristae morphogenesis. Biochim. Biophys. Acta Mol. Cell Res. 2009, 1793, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Nijtmans, L.G.J.; de Jong, L.; Artal Sanz, M.; Coates, P.J.; Berden, J.A.; Back, J.W.; Muijsers, A.O.; van der Spek, H.; Grivell, L.A. Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 2000, 19, 2444–2451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kehrein, K.; Schilling, R.; Vargas Möller-Hergt, B.; Wurm, C.A.; Jakobs, S.; Lamkemeyer, T.; Langer, T.; Ott, M. Organization of mitochondrial gene expression in two distinct ribosome-containing assemblies. Cell Rep. 2015, 10, 843–853. [Google Scholar] [CrossRef] [PubMed]
- Vargas Möller-Hergt, B.; Carlström, A.; Stephan, K.; Imhof, A.; Ott, M. The ribosome receptors Mrx15 and Mba1 jointly organize co-translational insertion and protein biogenesis in mitochondria. Mol. Biol. Cell 2018, 29, 2386–2396. [Google Scholar] [CrossRef] [PubMed]
- Preuss, M.; Leonhard, K.; Hell, K.; Stuart, R.A.; Neupert, W.; Herrmann, J.M. Mba1, a novel component of the mitochondrial protein export machinery of the yeast Saccharomyces cerevisiae. J. Cell Biol. 2001, 153, 1085–1096. [Google Scholar] [CrossRef] [PubMed]
- Ott, M.; Prestele, M.; Bauerschmitt, H.; Funes, S.; Bonnefoy, N.; Herrmann, J.M. Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J. 2006, 25, 1603–1610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfeffer, S.; Woellhaf, M.W.; Herrmann, J.M.; Förster, F. Organization of the mitochondrial translation machinery studied in situ by cryoelectron tomography. Nat. Commun. 2015, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bauerschmitt, H.; Mick, D.U.; Deckers, M.; Vollmer, C.; Funes, S.; Kehrein, K.; Ott, M.; Rehling, P.; Herrmann, J.M. Ribosome-binding proteins Mdm38 and Mba1 display overlapping functions for regulation of mitochondrial translation. Mol. Biol. Cell 2010, 21, 1937–1944. [Google Scholar] [CrossRef] [PubMed]
- Gruschke, S.; Kehrein, K.; Rompler, K.; Grone, K.; Israel, L.; Imhof, A.; Herrmann, J.M.; Ott, M. Cbp3-Cbp6 interacts with the yeast mitochondrial ribosomal tunnel exit and promotes cytochrome b synthesis and assembly. J. Cell Biol. 2011, 193, 1101–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, D.N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 2014, 12, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Suhm, T.; Kaimal, J.M.; Dawitz, H.; Peselj, C.; Masser, A.E.; Hanzén, S.; Ambrožič, M.; Smialowska, A.; Björck, M.L.; Brzezinski, P.; et al. Mitochondrial translation efficiency controls cytoplasmic protein homeostasis. Cell Metab. 2018, 27, 1309–1322. [Google Scholar] [CrossRef] [PubMed]
- Umeda, N.; Suzuki, T.; Yukawa, M.; Ohya, Y.; Shindo, H.; Watanabe, K.; Suzuki, T. Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs: Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem. 2005, 280, 1613–1624. [Google Scholar] [CrossRef] [PubMed]
- Colby, G.; Wu, M.; Tzagoloff, A. MTO1 codes for a mitochondrial protein required for respiration in paromomycin-resistant mutants of Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 27945–27952. [Google Scholar] [CrossRef] [PubMed]
- Hallstrom, T.C.; Moye-Rowley, W.S. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 37347–37356. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Li, L.; Jiang, H. Mitochondrial inner-membrane protease Yme1 degrades outer-membrane proteins Tom22 and Om45. J. Cell Biol. 2018, 217, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Graef, M.; Seewald, G.; Langer, T. Substrate recognition by AAA+ ATPases: Distinct substrate binding modes in ATP-dependent protease Yme1 of the mitochondrial intermembrane space. Mol. Cell. Biol. 2007, 27, 2476–2485. [Google Scholar] [CrossRef] [PubMed]
- Pearce, D.A.; Sherman, F. Degradation of cytochrome oxidase subunits in mutants of yeast lacking cytochrome c and suppression of the degradation by mutation of Yme1. J. Biol. Chem. 1995, 270, 20879–20882. [Google Scholar] [CrossRef] [PubMed]
- Nakai, T.; Yasuhara, T.; Fujiki, Y.; Ohashi, A. Multiple genes, including a member of the AAA family, are essential for degradation of unassembled subunit 2 of cytochrome c oxidase in yeast mitochondria. Mol. Cell. Biol. 1995, 15, 4441–4452. [Google Scholar] [CrossRef] [PubMed]
- Rep, M.; Nooy, J.; Guélin, E.; Grivell, L.A. Three genes for mitochondrial proteins suppress null-mutations in both Afg3 and Rca1 when over expressed. Curr. Genet. 1996, 30, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Van Bloois, E.; Dekker, H.L.; Fröderberg, L.; Houben, E.N.G.; Urbanus, M.L.; de Koster, C.G.; de Gier, J.W.; Luirink, J. Detection of cross-links between FtsH, YidC, HflK/C suggests a linked role for these proteins in quality control upon insertion of bacterial inner membrane proteins. FEBS Lett. 2008, 582, 1419–1424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preuss, M.; Ott, M.; Funes, S.; Luirink, J.; Herrmann, J.M. Evolution of mitochondrial oxa proteins from bacterial YidC: Inherited and acquired functions of a conserved protein insertion machinery. J. Biol. Chem. 2005, 280, 13004–13011. [Google Scholar] [CrossRef] [PubMed]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vargas Möller-Hergt, B.; Carlström, A.; Suhm, T.; Ott, M. Insertion Defects of Mitochondrially Encoded Proteins Burden the Mitochondrial Quality Control System. Cells 2018, 7, 172. https://doi.org/10.3390/cells7100172
Vargas Möller-Hergt B, Carlström A, Suhm T, Ott M. Insertion Defects of Mitochondrially Encoded Proteins Burden the Mitochondrial Quality Control System. Cells. 2018; 7(10):172. https://doi.org/10.3390/cells7100172
Chicago/Turabian StyleVargas Möller-Hergt, Braulio, Andreas Carlström, Tamara Suhm, and Martin Ott. 2018. "Insertion Defects of Mitochondrially Encoded Proteins Burden the Mitochondrial Quality Control System" Cells 7, no. 10: 172. https://doi.org/10.3390/cells7100172