Progesterone Receptor Membrane Component 1 (PGRMC1) Modulates Tumour Progression, the Immune Microenvironment and the Response to Therapy in Glioblastoma
Abstract
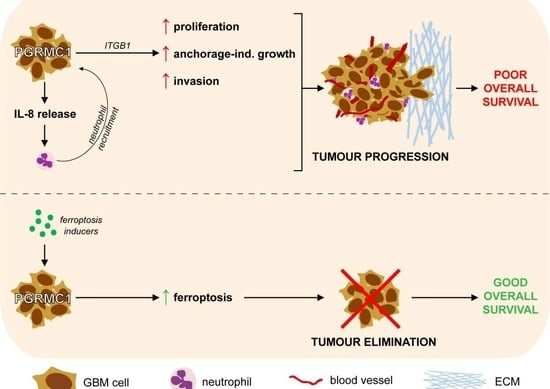
:1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Tissue Microarrays (TMA): Immunohistochemistry and Scoring
2.3. Cells and Supernatants
2.4. SDS-PAGE and Western Blot
2.5. Gene Expression Analysis
2.6. MTT Assay
2.7. Soft Agar Clonogenic Assay
2.8. Invasion Assay
2.9. Cell Death/Survival Assay
2.10. Chemotaxis Assay
2.11. MMP9 Release
2.12. Interleukin-8 Release
2.13. Statistical Analysis
3. Results
3.1. PGRMC1 Expression and Clinical Relevance in Glioblastoma (GBM) Patients
3.2. PGRMC1 and GBM Tumour Cell Functions
3.3. Molecular Mechanisms of PGRMC1 in GBM
3.4. PGRMC1 and GBM Therapy
3.5. PGRMC1 and the Immune Microenvironment in GBM
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012–2016. Neuro Oncol. 2019, 21, v1–v100. [Google Scholar] [CrossRef]
- Tan, A.C.; Ashley, D.M.; Lopez, G.Y.; Malinzak, M.; Friedman, H.S.; Khasraw, M. Management of glioblastoma: State of the art and future directions. CA Cancer J. Clin. 2020, 70, 299–312. [Google Scholar] [CrossRef]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.Y.; Weller, M.; Lee, E.Q.; Alexander, B.M.; Barnholtz-Sloan, J.S.; Barthel, F.P.; Batchelor, T.T.; Bindra, R.S.; Chang, S.M.; Chiocca, E.A.; et al. Glioblastoma in adults: A Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) consensus review on current management and future directions. Neuro Oncol. 2020, 22, 1073–1113. [Google Scholar] [CrossRef]
- Cruz Da Silva, E.; Mercier, M.C.; Etienne-Selloum, N.; Dontenwill, M.; Choulier, L. A Systematic Review of Glioblastoma-Targeted Therapies in Phases II, III, IV Clinical Trials. Cancers 2021, 13, 1795. [Google Scholar] [CrossRef]
- Cahill, M.A.; Jazayeri, J.A.; Catalano, S.M.; Toyokuni, S.; Kovacevic, Z.; Richardson, D.R. The emerging role of progesterone receptor membrane component 1 (PGRMC1) in cancer biology. Biochim. Biophys. Acta 2016, 1866, 339–349. [Google Scholar] [CrossRef]
- Cahill, M.A.; Neubauer, H. PGRMC Proteins Are Coming of Age: A Special Issue on the Role of PGRMC1 and PGRMC2 in Metabolism and Cancer Biology. Cancers 2021, 13, 512. [Google Scholar] [CrossRef]
- Lee, S.R.; Lee, J.G.; Heo, J.H.; Jo, S.L.; Ryu, J.; Kim, G.; Yon, J.M.; Lee, M.S.; Lee, G.S.; An, B.S.; et al. Loss of PGRMC1 Delays the Progression of Hepatocellular Carcinoma via Suppression of Pro-Inflammatory Immune Responses. Cancers 2021, 13, 2438. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Ruan, X.; Zhang, Y.; Cai, G.; Ju, R.; Yang, Y.; Cheng, J.; Gu, M. Comprehensive Analysis of the Implication of PGRMC1 in Triple-Negative Breast Cancer. Front. Bioeng. Biotechnol. 2021, 9, 714030. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Xia, X.; Wang, X.; Zhang, P.; Lu, W.; Yu, Y.; Deng, S.; Yang, H.; Zhu, H.; Xu, N.; et al. PGRMC1 Is a Novel Potential Tumor Biomarker of Human Renal Cell Carcinoma Based on Quantitative Proteomic and Integrative Biological Assessments. PLoS ONE 2017, 12, e0170453. [Google Scholar] [CrossRef]
- Zhao, Y.; Ruan, X. Identification of PGRMC1 as a Candidate Oncogene for Head and Neck Cancers and Its Involvement in Metabolic Activities. Front. Bioeng. Biotechnol. 2019, 7, 438. [Google Scholar] [CrossRef] [PubMed]
- Clark, N.C.; Friel, A.M.; Pru, C.A.; Zhang, L.; Shioda, T.; Rueda, B.R.; Peluso, J.J.; Pru, J.K. Progesterone receptor membrane component 1 promotes survival of human breast cancer cells and the growth of xenograft tumors. Cancer Biol. Ther. 2016, 17, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Friel, A.M.; Zhang, L.; Pru, C.A.; Clark, N.C.; McCallum, M.L.; Blok, L.J.; Shioda, T.; Peluso, J.J.; Rueda, B.R.; Pru, J.K. Progesterone receptor membrane component 1 deficiency attenuates growth while promoting chemosensitivity of human endometrial xenograft tumors. Cancer Lett. 2015, 356, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.T.; May, E.W.; Chang, J.F.; Hu, R.Y.; Wang, L.H.; Chan, H.L. PGRMC1 contributes to doxorubicin-induced chemoresistance in MES-SA uterine sarcoma. Cell. Mol. Life Sci. 2015, 72, 2395–2409. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Higashisaka, K.; Shintani, T.; Maki, A.; Hanamuro, S.; Haga, Y.; Maeda, S.; Tsujino, H.; Nagano, K.; Fujio, Y.; et al. Progesterone receptor membrane component 1 leads to erlotinib resistance, initiating crosstalk of Wnt/beta-catenin and NF-kappaB pathways, in lung adenocarcinoma cells. Sci. Rep. 2020, 10, 4748. [Google Scholar] [CrossRef]
- Peluso, J.J.; Liu, X.; Saunders, M.M.; Claffey, K.P.; Phoenix, K. Regulation of ovarian cancer cell viability and sensitivity to cisplatin by progesterone receptor membrane component-1. J. Clin. Endocrinol. Metab. 2008, 93, 1592–1599. [Google Scholar] [CrossRef]
- Kabe, Y.; Nakane, T.; Koike, I.; Yamamoto, T.; Sugiura, Y.; Harada, E.; Sugase, K.; Shimamura, T.; Ohmura, M.; Muraoka, K.; et al. Haem-dependent dimerization of PGRMC1/Sigma-2 receptor facilitates cancer proliferation and chemoresistance. Nat. Commun. 2016, 7, 11030. [Google Scholar] [CrossRef]
- You, J.H.; Lee, J.; Roh, J.L. PGRMC1-dependent lipophagy promotes ferroptosis in paclitaxel-tolerant persister cancer cells. J. Exp. Clin. Cancer Res. 2021, 40, 350. [Google Scholar] [CrossRef]
- De Leo, A.; Ugolini, A.; Veglia, F. Myeloid Cells in Glioblastoma Microenvironment. Cells 2020, 10, 18. [Google Scholar] [CrossRef]
- Kresse, N.; Schroder, H.; Stein, K.P.; Wilkens, L.; Mawrin, C.; Sandalcioglu, I.E.; Dumitru, C.A. PLOD2 Is a Prognostic Marker in Glioblastoma That Modulates the Immune Microenvironment and Tumor Progression. Int. J. Mol. Sci. 2022, 23, 6037. [Google Scholar] [CrossRef]
- Zha, C.; Meng, X.; Li, L.; Mi, S.; Qian, D.; Li, Z.; Wu, P.; Hu, S.; Zhao, S.; Cai, J.; et al. Neutrophil extracellular traps mediate the crosstalk between glioma progression and the tumor microenvironment via the HMGB1/RAGE/IL-8 axis. Cancer Biol. Med. 2020, 17, 154–168. [Google Scholar] [CrossRef] [PubMed]
- Dumitru, C.A.; Brouwer, E.; Stelzer, T.; Nocerino, S.; Rading, S.; Wilkens, L.; Sandalcioglu, I.E.; Karsak, M. Dynein Light Chain Protein Tctex1: A Novel Prognostic Marker and Molecular Mediator in Glioblastoma. Cancers 2021, 13, 2624. [Google Scholar] [CrossRef] [PubMed]
- Schiffgens, S.; Wilkens, L.; Brandes, A.A.; Meier, T.; Franceschi, E.; Ermani, M.; Hartmann, C.; Sandalcioglu, I.E.; Dumitru, C.A. Sex-specific clinicopathological significance of novel (Frizzled-7) and established (MGMT, IDH1) biomarkers in glioblastoma. Oncotarget 2016, 7, 55169–55180. [Google Scholar] [CrossRef]
- Chambless, L.B.; Kistka, H.M.; Parker, S.L.; Hassam-Malani, L.; McGirt, M.J.; Thompson, R.C. The relative value of postoperative versus preoperative Karnofsky Performance Scale scores as a predictor of survival after surgical resection of glioblastoma multiforme. J. Neurooncol. 2015, 121, 359–364. [Google Scholar] [CrossRef]
- Filippini, G.; Falcone, C.; Boiardi, A.; Broggi, G.; Bruzzone, M.G.; Caldiroli, D.; Farina, R.; Farinotti, M.; Fariselli, L.; Finocchiaro, G.; et al. Prognostic factors for survival in 676 consecutive patients with newly diagnosed primary glioblastoma. Neuro Oncol. 2008, 10, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Liang, H.; Cheng, P.; Yang, H.; Zhao, P. Gross Total vs. Subtotal Resection on Survival Outcomes in Elderly Patients With High-Grade Glioma: A Systematic Review and Meta-Analysis. Front. Oncol. 2020, 10, 151. [Google Scholar] [CrossRef]
- Siker, M.L.; Wang, M.; Porter, K.; Nelson, D.F.; Curran, W.J.; Michalski, J.M.; Souhami, L.; Chakravarti, A.; Yung, W.K.; Delrowe, J.; et al. Age as an independent prognostic factor in patients with glioblastoma: A Radiation Therapy Oncology Group and American College of Surgeons National Cancer Data Base comparison. J. Neurooncol. 2011, 104, 351–356. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, J.K.; Jeon, H.Y.; Ham, S.W.; Kim, H. CD133 Regulates IL-1beta Signaling and Neutrophil Recruitment in Glioblastoma. Mol. Cells 2017, 40, 515–522. [Google Scholar] [CrossRef]
- Liang, J.; Piao, Y.; Holmes, L.; Fuller, G.N.; Henry, V.; Tiao, N.; de Groot, J.F. Neutrophils promote the malignant glioma phenotype through S100A4. Clin. Cancer Res. 2014, 20, 187–198. [Google Scholar] [CrossRef]
- Pedroza, D.A.; Rajamanickam, V.; Subramani, R.; Bencomo, A.; Galvez, A.; Lakshmanaswamy, R. Progesterone receptor membrane component 1 promotes the growth of breast cancers by altering the phosphoproteome and augmenting EGFR/PI3K/AKT signalling. Br. J. Cancer 2020, 123, 1326–1335. [Google Scholar] [CrossRef]
- Asperger, H.; Stamm, N.; Gierke, B.; Pawlak, M.; Hofmann, U.; Zanger, U.M.; Marton, A.; Katona, R.L.; Buhala, A.; Vizler, C.; et al. Progesterone receptor membrane component 1 regulates lipid homeostasis and drives oncogenic signaling resulting in breast cancer progression. Breast Cancer Res. 2020, 22, 75. [Google Scholar] [CrossRef]
- Huang, H.Y.; Chou, H.C.; Law, C.H.; Chang, W.T.; Wen, T.N.; Liao, E.C.; Lin, M.W.; Lin, L.H.; Wei, Y.S.; Tsai, Y.T.; et al. Progesterone receptor membrane component 1 is involved in oral cancer cell metastasis. J. Cell Mol. Med. 2020, 24, 9737–9751. [Google Scholar] [CrossRef] [PubMed]
- Blandin, A.F.; Renner, G.; Lehmann, M.; Lelong-Rebel, I.; Martin, S.; Dontenwill, M. beta1 Integrins as Therapeutic Targets to Disrupt Hallmarks of Cancer. Front. Pharmacol. 2015, 6, 279. [Google Scholar] [CrossRef] [PubMed]
- Ellert-Miklaszewska, A.; Poleszak, K.; Pasierbinska, M.; Kaminska, B. Integrin Signaling in Glioma Pathogenesis: From Biology to Therapy. Int. J. Mol. Sci. 2020, 21, 888. [Google Scholar] [CrossRef] [PubMed]
- Crudden, G.; Chitti, R.E.; Craven, R.J. Hpr6 (heme-1 domain protein) regulates the susceptibility of cancer cells to chemotherapeutic drugs. J. Pharmacol. Exp. Ther. 2006, 316, 448–455. [Google Scholar] [CrossRef]
- Wang, H.; Lin, D.; Yu, Q.; Li, Z.; Lenahan, C.; Dong, Y.; Wei, Q.; Shao, A. A Promising Future of Ferroptosis in Tumor Therapy. Front. Cell Dev. Biol. 2021, 9, 629150. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dumitru, C.A.; Schröder, H.; Schäfer, F.T.A.; Aust, J.F.; Kreße, N.; Siebert, C.L.R.; Stein, K.-P.; Haghikia, A.; Wilkens, L.; Mawrin, C.; et al. Progesterone Receptor Membrane Component 1 (PGRMC1) Modulates Tumour Progression, the Immune Microenvironment and the Response to Therapy in Glioblastoma. Cells 2023, 12, 2498. https://doi.org/10.3390/cells12202498
Dumitru CA, Schröder H, Schäfer FTA, Aust JF, Kreße N, Siebert CLR, Stein K-P, Haghikia A, Wilkens L, Mawrin C, et al. Progesterone Receptor Membrane Component 1 (PGRMC1) Modulates Tumour Progression, the Immune Microenvironment and the Response to Therapy in Glioblastoma. Cells. 2023; 12(20):2498. https://doi.org/10.3390/cells12202498
Chicago/Turabian StyleDumitru, Claudia Alexandra, Hannah Schröder, Frederik Till Alexander Schäfer, Jan Friedrich Aust, Nina Kreße, Carl Ludwig Raven Siebert, Klaus-Peter Stein, Aiden Haghikia, Ludwig Wilkens, Christian Mawrin, and et al. 2023. "Progesterone Receptor Membrane Component 1 (PGRMC1) Modulates Tumour Progression, the Immune Microenvironment and the Response to Therapy in Glioblastoma" Cells 12, no. 20: 2498. https://doi.org/10.3390/cells12202498