Spaceflight Induces Strength Decline in Caenorhabditis elegans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Worm Preparation
2.2. Fabrication and Assembly of a Microfluidic Device in Worm Loading Apparatus (WLA)
2.3. Image Acquisition and Processing
2.4. RNA Extraction, Sequencing and Data Pre-Processing
2.5. Gene Expression Analysis
3. Results
3.1. Growth
3.2. Body Diameter and Length
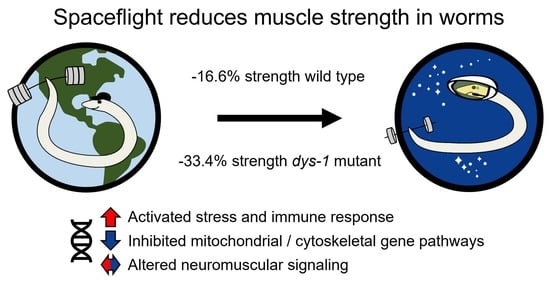
3.3. Muscle Strength
3.4. Gene Expression Analysis
3.5. Genes Predicted to Be Altered in Space Are Predicted to Be Altered by Drugs on Earth
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koehle, A.P.; Brumwell, S.L.; Seto, E.P.; Lynch, A.M.; Urbaniak, C. Microbial Applications for Sustainable Space Exploration beyond Low Earth Orbit. npj Microgravity 2023, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Patel, Z.S.; Brunstetter, T.J.; Tarver, W.J.; Whitmire, A.M.; Zwart, S.R.; Smith, S.M.; Huff, J.L. Red Risks for a Journey to the Red Planet: The Highest Priority Human Health Risks for a Mission to Mars. npj Microgravity 2020, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, J.; Hattori, A.; Takahashi, A.; Furusawa, Y.; Tabuchi, Y.; Shibata, M.; Nagamatsu, A.; Yano, S.; Maruyama, Y.; Matsubara, H.; et al. Physiological Consequences of Space Flight, Including Abnormal Bone Metabolism, Space Radiation Injury, and Circadian Clock Dysregulation: Implications of Melatonin Use and Regulation as a Countermeasure. J. Pineal Res. 2023, 74, e12834. [Google Scholar] [CrossRef]
- Sy, M.R.; Keefe, J.A.; Sutton, J.P.; Wehrens, X.H.T. Cardiac Function, Structural, and Electrical Remodeling by Microgravity Exposure. Am. J. Physiol. Heart Circ. Physiol. 2023, 324, H1–H13. [Google Scholar] [CrossRef] [PubMed]
- Ong, J.; Tarver, W.; Brunstetter, T.; Mader, T.H.; Gibson, C.R.; Mason, S.S.; Lee, A. Spaceflight Associated Neuro-Ocular Syndrome: Proposed Pathogenesis, Terrestrial Analogues, and Emerging Countermeasures. Br. J. Ophthalmol. 2023, 107, 895–900. [Google Scholar] [CrossRef]
- Liphardt, A.-M.; Fernandez-Gonzalo, R.; Albracht, K.; Rittweger, J.; Vico, L. Musculoskeletal Research in Human Space Flight—Unmet Needs for the Success of Crewed Deep Space Exploration. npj Microgravity 2023, 9, 9. [Google Scholar] [CrossRef]
- Stahn, A.C.; Bucher, D.; Zu Eulenburg, P.; Denise, P.; Smith, N.; Pagnini, F.; White, O. Paving the Way to Better Understand the Effects of Prolonged Spaceflight on Operational Performance and Its Neural Bases. npj Microgravity 2023, 9, 59. [Google Scholar] [CrossRef]
- Winkler, L.H. Human Physiological Limitations to Long-Term Spaceflight and Living in Space. Aerosp. Med. Hum. Perform. 2023, 94, 444–456. [Google Scholar] [CrossRef]
- Krittanawong, C.; Singh, N.K.; Scheuring, R.A.; Urquieta, E.; Bershad, E.M.; Macaulay, T.R.; Kaplin, S.; Dunn, C.; Kry, S.F.; Russomano, T.; et al. Human Health during Space Travel: State-of-the-Art Review. Cells 2022, 12, 40. [Google Scholar] [CrossRef]
- Lane, H.W.; Bourland, C.; Barrett, A.; Heer, M.; Smith, S.M. The Role of Nutritional Research in the Success of Human Space Flight. Adv. Nutr. 2013, 4, 521–523. [Google Scholar] [CrossRef]
- Roy-O’Reilly, M.; Mulavara, A.; Williams, T. A Review of Alterations to the Brain during Spaceflight and the Potential Relevance to Crew in Long-Duration Space Exploration. npj Microgravity 2021, 7, 5. [Google Scholar] [CrossRef]
- Afshinnekoo, E.; Scott, R.T.; MacKay, M.J.; Pariset, E.; Cekanaviciute, E.; Barker, R.; Gilroy, S.; Hassane, D.; Smith, S.M.; Zwart, S.R.; et al. Fundamental Biological Features of Spaceflight: Advancing the Field to Enable Deep-Space Exploration. Cell 2020, 183, 1162–1184. [Google Scholar] [CrossRef]
- Thirsk, R.; Kuipers, A.; Mukai, C.; Williams, D. The Space-Flight Environment: The International Space Station and beyond. CMAJ 2009, 180, 1216–1220. [Google Scholar] [CrossRef] [PubMed]
- Fogtman, A.; Baatout, S.; Baselet, B.; Berger, T.; Hellweg, C.E.; Jiggens, P.; La Tessa, C.; Narici, L.; Nieminen, P.; Sabatier, L.; et al. Towards Sustainable Human Space Exploration-Priorities for Radiation Research to Quantify and Mitigate Radiation Risks. npj Microgravity 2023, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Braun, N.; Thomas, S.; Tronnier, H.; Heinrich, U. Self-Reported Skin Changes by a Selected Number of Astronauts after Long-Duration Mission on ISS as Part of the Skin B Project. Skin Pharmacol. Physiol. 2019, 32, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Pavez Loriè, E.; Baatout, S.; Choukér, A.; Buchheim, J.-I.; Baselet, B.; Dello Russo, C.; Wotring, V.; Monici, M.; Morbidelli, L.; Gagliardi, D.; et al. The Future of Personalized Medicine in Space: From Observations to Countermeasures. Front. Bioeng. Biotechnol. 2021, 9, 739747. [Google Scholar] [CrossRef]
- Stroud, J.E.; Gale, M.S.; Zwart, S.R.; Heer, M.; Smith, S.M.; Montina, T.; Metz, G.A.S. Longitudinal Metabolomic Profiles Reveal Sex-Specific Adjustments to Long-Duration Spaceflight and Return to Earth. Cell. Mol. Life Sci. 2022, 79, 578. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.T.; Sanders, L.M.; Antonsen, E.L.; Hastings, J.J.A.; Park, S.-M.; Mackintosh, G.; Reynolds, R.J.; Hoarfrost, A.L.; Sawyer, A.; Greene, C.S.; et al. Biomonitoring and Precision Health in Deep Space Supported by Artificial Intelligence. Nat. Mach. Intell. 2023, 5, 196–207. [Google Scholar] [CrossRef]
- Griko, Y.V.; Loftus, D.J.; Stolc, V.; Peletskaya, E. Private Spaceflight: A New Landscape for Dealing with Medical Risk. Life Sci. Space Res. 2022, 33, 41–47. [Google Scholar] [CrossRef]
- Straume, T.; Loftus, D.; Li, J.; Coleman, M.; Davis, C.; McMonigal, K.; Piccini, M.; Singh, A. Biomarker-Detection Technologies for Comprehensive Medical Diagnosis during Deep-Space Missions. Recent Pat. Space Technol. 2013, 3, 13–23. [Google Scholar] [CrossRef]
- Garrett-Bakelman, F.E.; Darshi, M.; Green, S.J.; Gur, R.C.; Lin, L.; Macias, B.R.; McKenna, M.J.; Meydan, C.; Mishra, T.; Nasrini, J.; et al. The NASA Twins Study: A Multidimensional Analysis of a Year-Long Human Spaceflight. Science 2019, 364, eaau8650. [Google Scholar] [CrossRef] [PubMed]
- Cope, H.; Willis, C.R.G.; MacKay, M.J.; Rutter, L.A.; Toh, L.S.; Williams, P.M.; Herranz, R.; Borg, J.; Bezdan, D.; Giacomello, S.; et al. Routine Omics Collection Is a Golden Opportunity for European Human Research in Space and Analog Environments. Patterns 2022, 3, 100550. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Charvat, J.; Zwart, S.R.; Mehta, S.K.; Crucian, B.E.; Smith, S.M.; He, J.; Piermarocchi, C.; Mias, G.I. Time-Resolved Molecular Measurements Reveal Changes in Astronauts during Spaceflight. Front. Physiol. 2023, 14, 1219221. [Google Scholar] [CrossRef] [PubMed]
- Palmer, R.H.C.; Johnson, E.C.; Won, H.; Polimanti, R.; Kapoor, M.; Chitre, A.; Bogue, M.A.; Benca-Bachman, C.E.; Parker, C.C.; Verma, A.; et al. Integration of Evidence across Human and Model Organism Studies: A Meeting Report. Genes Brain Behav. 2021, 20, e12738. [Google Scholar] [CrossRef]
- Yamamoto, S.; Kanca, O.; Wangler, M.F.; Bellen, H.J. Integrating Non-Mammalian Model Organisms in the Diagnosis of Rare Genetic Diseases in Humans. Nat. Rev. Genet. 2023. [Google Scholar] [CrossRef] [PubMed]
- Bult, C.J.; Sternberg, P.W. The Alliance of Genome Resources: Transforming Comparative Genomics. Mamm. Genome 2023. [Google Scholar] [CrossRef]
- Rutter, L.; Barker, R.; Bezdan, D.; Cope, H.; Costes, S.V.; Degoricija, L.; Fisch, K.M.; Gabitto, M.I.; Gebre, S.; Giacomello, S.; et al. A New Era for Space Life Science: International Standards for Space Omics Processing. Patterns 2020, 1, 100148. [Google Scholar] [CrossRef]
- Mathyk, B.A.; Tabetah, M.; Karim, R.; Zaksas, V.; Kim, J.; Anu, I.; Muratani, M.; Tasoula, A.; Singh, R.; Chen, Y.-K.; et al. Spaceflight Alters Insulin and Estrogen Signaling Pathways. Res. Sq. 2023. [Google Scholar] [CrossRef]
- Akintola, A.A.; van Heemst, D. Insulin, Aging, and the Brain: Mechanisms and Implications. Front. Endocrinol. 2015, 6, 13. [Google Scholar] [CrossRef]
- Vitry, G.; Finch, R.; Mcstay, G.; Behesti, A.; Déjean, S.; Larose, T.; Wotring, V.; da Silveira, W.A. Muscle Atrophy Phenotype Gene Expression during Spaceflight Is Linked to a Metabolic Crosstalk in Both the Liver and the Muscle in Mice. iScience 2022, 25, 105213. [Google Scholar] [CrossRef]
- Institute of Medicine (US) Committee on Creating a Vision for Space Medicine during Travel Beyond Earth Orbit; Ball, J.R.; Evans, C.H., Jr. Astronaut Health Beyond Earth Orbit; National Academies Press: Washington, DC, USA, 2001. [Google Scholar]
- Laws, J.M.; Caplan, N.; Bruce, C.; McGrogan, C.; Lindsay, K.; Wild, B.; Debuse, D.; Wotring, V.; Winnard, A. Systematic Review of the Technical and Physiological Constraints of the Orion Multi-Purpose Crew Vehicle That Affect the Capability of Astronauts to Exercise Effectively during Spaceflight. Acta Astronaut. 2020, 170, 665–677. [Google Scholar] [CrossRef]
- Tang, H.; Rising, H.H.; Majji, M.; Brown, R.D. Long-Term Space Nutrition: A Scoping Review. Nutrients 2021, 14, 094. [Google Scholar] [CrossRef] [PubMed]
- Kumar, L.; Gaikwad, K.K. Advanced Food Packaging Systems for Space Exploration Missions. Life Sci. Space Res. 2023, 37, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Pandith, J.A.; Neekhra, S.; Ahmad, S.; Sheikh, R.A. Recent Developments in Space Food for Exploration Missions: A Review. Life Sci. Space Res. 2023, 36, 123–134. [Google Scholar] [CrossRef]
- Craig Everroad, R.; Foster, J.; Galazka, J.M.; Jansson, J.; Lee, J.A.; Lera, M.P.; Perera, I.; Ricco, A.; Szewczyk, N.; Todd, P.; et al. Space Biology Beyond LEO Instrumentation & Science Series—Science Working Group 2021 Annual Report; National Aeronautics and Space Administration: Washington, DC, USA, 2021. [Google Scholar]
- Sanders, L.M.; Scott, R.T.; Yang, J.H.; Qutub, A.A.; Garcia Martin, H.; Berrios, D.C.; Hastings, J.J.A.; Rask, J.; Mackintosh, G.; Hoarfrost, A.L.; et al. Biological Research and Self-Driving Labs in Deep Space Supported by Artificial Intelligence. Nat. Mach. Intell. 2023, 5, 208–219. [Google Scholar] [CrossRef]
- Elsaesser, A.; Burr, D.J.; Mabey, P.; Urso, R.G.; Billi, D.; Cockell, C.; Cottin, H.; Kish, A.; Leys, N.; van Loon, J.J.W.A.; et al. Future Space Experiment Platforms for Astrobiology and Astrochemistry Research. npj Microgravity 2023, 9, 43. [Google Scholar] [CrossRef]
- Massaro Tieze, S.; Liddell, L.C.; Santa Maria, S.R.; Bhattacharya, S. BioSentinel: A Biological CubeSat for Deep Space Exploration. Astrobiology 2023, 23, 631–636. [Google Scholar] [CrossRef]
- Zea, L.; Piper, S.S.; Gaikani, H.; Khoshnoodi, M.; Niederwieser, T.; Hoehn, A.; Grusin, M.; Wright, J.; Flores, P.; Wilson, K.; et al. Experiment Verification Test of the Artemis I “Deep Space Radiation Genomics” Experiment. Acta Astronaut. 2022, 198, 702–706. [Google Scholar] [CrossRef]
- National Academies of Sciences, Engineering, and Medicine. Report Series: Committee on Biological and Physical Sciences in Space: Using Commercial Lunar Payload Services (CLPS) to Achieve Lunar Biological and Physical Science Objectives: Proceedings of a Workshop; The National Academies Press: Washington, DC, USA, 2021. [Google Scholar]
- Rahman, M.; Hewitt, J.E.; Van-Bussel, F.; Edwards, H.; Blawzdziewicz, J.; Szewczyk, N.J.; Driscoll, M.; Vanapalli, S.A. NemaFlex: A Microfluidics-Based Technology for Standardized Measurement of Muscular Strength of C. elegans. Lab Chip 2018, 18, 2187–2201. [Google Scholar] [CrossRef]
- Lesanpezeshki, L.; Qadota, H.; Darabad, M.N.; Kashyap, K.; Lacerda, C.M.R.; Szewczyk, N.J.; Benian, G.M.; Vanapalli, S.A. Investigating the Correlation of Muscle Function Tests and Sarcomere Organization in C. elegans. Skelet. Muscle 2021, 11, 20. [Google Scholar] [CrossRef]
- Hewitt, J.E.; Pollard, A.K.; Lesanpezeshki, L.; Deane, C.S.; Gaffney, C.J.; Etheridge, T.; Szewczyk, N.J.; Vanapalli, S.A. Muscle Strength Deficiency and Mitochondrial Dysfunction in a Muscular Dystrophy Model of Caenorhabditis elegans and Its Functional Response to Drugs. Dis. Model. Mech. 2018, 11, dmm036137. [Google Scholar] [CrossRef] [PubMed]
- Etheridge, T.; Rahman, M.; Gaffney, C.J.; Shaw, D.; Shephard, F.; Magudia, J.; Solomon, D.E.; Milne, T.; Blawzdziewicz, J.; Constantin-Teodosiu, D.; et al. The Integrin-Adhesome Is Required to Maintain Muscle Structure, Mitochondrial ATP Production, and Movement Forces in Caenorhabditis elegans. FASEB J. 2015, 29, 1235–1246. [Google Scholar] [CrossRef] [PubMed]
- Ellwood, R.A.; Hewitt, J.E.; Torregrossa, R.; Philp, A.M.; Hardee, J.P.; Hughes, S.; van de Klashorst, D.; Gharahdaghi, N.; Anupom, T.; Slade, L.; et al. Mitochondrial Hydrogen Sulfide Supplementation Improves Health in the C. elegans Duchenne Muscular Dystrophy Model. Proc. Natl. Acad. Sci. USA 2021, 118, e2018342118. [Google Scholar] [CrossRef] [PubMed]
- Vintila, A.R.; Slade, L.; Cooke, M.; Willis, C.R.G.; Torregrossa, R.; Rahman, M.; Anupom, T.; Vanapalli, S.A.; Gaffney, C.J.; Gharahdaghi, N.; et al. Mitochondrial Sulfide Promotes Life Span and Health Span through Distinct Mechanisms in Developing versus Adult Treated Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2023, 120, e2216141120. [Google Scholar] [CrossRef]
- Scott, A.; Willis, C.R.G.; Muratani, M.; Higashitani, A.; Etheridge, T.; Szewczyk, N.J.; Deane, C.S. Caenorhabditis elegans in Microgravity: An Omics Perspective. iScience 2023, 26, 107189. [Google Scholar] [CrossRef]
- Soni, P.; Anupom, T.; Lesanpezeshki, L.; Rahman, M.; Hewitt, J.E.; Vellone, M.; Stodieck, L.; Blawzdziewicz, J.; Szewczyk, N.J.; Vanapalli, S.A. Microfluidics-Integrated Spaceflight Hardware for Measuring Muscle Strength of Caenorhabditis elegans on the International Space Station. npj Microgravity 2022, 8, 50. [Google Scholar] [CrossRef]
- Ishioka, N.; Higashibata, A. Space Experiments Using C. elegans as a Model Organism. In Handbook of Space Pharmaceuticals; Pathak, Y.V., Araújo dos Santos, M., Zea, L., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 647–677. ISBN 9783030055264. [Google Scholar]
- Higashibata, A.; Szewczyk, N.J.; Conley, C.A.; Imamizo-Sato, M.; Higashitani, A.; Ishioka, N. Decreased Expression of Myogenic Transcription Factors and Myosin Heavy Chains in Caenorhabditis elegans Muscles Developed during Spaceflight. J. Exp. Biol. 2006, 209, 3209–3218. [Google Scholar] [CrossRef]
- Higashibata, A.; Hashizume, T.; Nemoto, K.; Higashitani, N.; Etheridge, T.; Mori, C.; Harada, S.; Sugimoto, T.; Szewczyk, N.J.; Baba, S.A.; et al. Microgravity Elicits Reproducible Alterations in Cytoskeletal and Metabolic Gene and Protein Expression in Space-Flown Caenorhabditis elegans. npj Microgravity 2016, 2, 15022. [Google Scholar] [CrossRef]
- Honda, Y.; Higashibata, A.; Matsunaga, Y.; Yonezawa, Y.; Kawano, T.; Higashitani, A.; Kuriyama, K.; Shimazu, T.; Tanaka, M.; Szewczyk, N.J.; et al. Genes down-Regulated in Spaceflight Are Involved in the Control of Longevity in Caenorhabditis elegans. Sci. Rep. 2012, 2, 487. [Google Scholar] [CrossRef]
- Sterken, M.G.; Snoek, L.B.; Kammenga, J.E.; Andersen, E.C. The Laboratory Domestication of Caenorhabditis elegans. Trends Genet. 2015, 31, 224–231. [Google Scholar] [CrossRef]
- Oh, K.H.; Kim, H. Reduced IGF Signaling Prevents Muscle Cell Death in a Caenorhabditis elegans Model of Muscular Dystrophy. Proc. Natl. Acad. Sci. USA 2013, 110, 19024–19029. [Google Scholar] [CrossRef]
- Szewczyk, N.J.; Kozak, E.; Conley, C.A. Chemically Defined Medium and Caenorhabditis elegans. BMC Biotechnol. 2003, 3, 19. [Google Scholar] [CrossRef]
- Zare, M.; Mikkonen, K.S. Phase Change Materials for Life Science Applications. Adv. Funct. Mater. 2023, 33, 2213455. [Google Scholar] [CrossRef]
- Choi, S.; Park, J.-K. Two-Step Photolithography to Fabricate Multilevel Microchannels. Biomicrofluidics 2010, 4, 46503. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce Acceleration-Supported Software for Integrated Quality Control and Preprocessing of High-Throughput Sequencing Data. Gigascience 2018, 7, gix120. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential Analyses for RNA-Seq: Transcript-Level Estimates Improve Gene-Level Inferences. F1000Research 2015, 4, 1521. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Stephens, M. False Discovery Rates: A New Deal. Biostatistics 2017, 18, 275–294. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Cary, M.; Podshivalova, K.; Kenyon, C. Application of Transcriptional Gene Modules to Analysis of Caenorhabditis elegans’ Gene Expression Data. G3 2020, 10, 3623–3638. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Underwood, R.S.; Greenwald, I.; Shaye, D.D. OrthoList 2: A New Comparative Genomic Analysis of Human and Caenorhabditis elegans Genes. Genetics 2018, 210, 445–461. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal Analysis Approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Oczypok, E.A.; Etheridge, T.; Freeman, J.; Stodieck, L.; Johnsen, R.; Baillie, D.; Szewczyk, N.J. Remote Automated Multi-Generational Growth and Observation of an Animal in Low Earth Orbit. J. R. Soc. Interface 2012, 9, 596–599. [Google Scholar] [CrossRef]
- Szewczyk, N.J.; Tillman, J.; Conley, C.A.; Granger, L.; Segalat, L.; Higashitani, A.; Honda, S.; Honda, Y.; Kagawa, H.; Adachi, R.; et al. Description of International Caenorhabditis elegans Experiment First Flight (ICE-FIRST). Adv. Space Res. 2008, 42, 1072–1079. [Google Scholar] [CrossRef]
- Nelson, G.A.; Schubert, W.W.; Kazarians, G.A.; Richards, G.F. Development and Chromosome Mechanics in Nematodes: Results from IML-1. Adv. Space Res. 1994, 14, 209–214. [Google Scholar] [CrossRef]
- Nelson, G.A.; Schubert, W.W.; Kazarians, G.A.; Richards, G.F.; Benton, E.V.; Benton, E.R.; Henke, R. Radiation Effects in Nematodes: Results from IML-1 Experiments. Adv. Space Res. 1994, 14, 87–91. [Google Scholar] [CrossRef]
- Qiao, L.; Luo, S.; Liu, Y.; Li, X.; Wang, G.; Huang, Z. Reproductive and Locomotory Capacities of Caenorhabditis elegans Were Not Affected by Simulated Variable Gravities and Spaceflight during the Shenzhou-8 Mission. Astrobiology 2013, 13, 617–625. [Google Scholar] [CrossRef]
- Zhao, Y.; Lai, K.; Cheung, I.; Youds, J.; Tarailo, M.; Tarailo, S.; Rose, A. A Mutational Analysis of Caenorhabditis elegans in Space. Mutat. Res. 2006, 601, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Sudevan, S.; Muto, K.; Higashitani, N.; Hashizume, T.; Higashibata, A.; Ellwood, R.A.; Deane, C.S.; Rahman, M.; Vanapalli, S.A.; Etheridge, T.; et al. Loss of Physical Contact in Space Alters the Dopamine System in C. elegans. iScience 2022, 25, 103762. [Google Scholar] [CrossRef] [PubMed]
- Selch, F.; Higashibata, A.; Imamizo-Sato, M.; Higashitani, A.; Ishioka, N.; Szewczyk, N.J.; Conley, C.A. Genomic Response of the Nematode Caenorhabditis elegans to Spaceflight. Adv. Space Res. 2008, 41, 807–815. [Google Scholar] [CrossRef]
- Harada, S.; Hashizume, T.; Nemoto, K.; Shao, Z.; Higashitani, N.; Etheridge, T.; Szewczyk, N.J.; Fukui, K.; Higashibata, A.; Higashitani, A. Fluid Dynamics Alter Caenorhabditis elegans Body Length via TGF-β/DBL-1 Neuromuscular Signaling. npj Microgravity 2016, 2, 16006. [Google Scholar] [CrossRef] [PubMed]
- Szewczyk, N.J.; Udranszky, I.A.; Kozak, E.; Sunga, J.; Kim, S.K.; Jacobson, L.A.; Conley, C.A. Delayed Development and Lifespan Extension as Features of Metabolic Lifestyle Alteration in C. elegans under Dietary Restriction. J. Exp. Biol. 2006, 209, 4129–4139. [Google Scholar] [CrossRef] [PubMed]
- English, K.L.; Downs, M.; Goetchius, E.; Buxton, R.; Ryder, J.W.; Ploutz-Snyder, R.; Guilliams, M.; Scott, J.M.; Ploutz-Snyder, L.L. High Intensity Training during Spaceflight: Results from the NASA Sprint Study. npj Microgravity 2020, 6, 21. [Google Scholar] [CrossRef]
- Xu, D.; Gao, Y.; Guo, L.; Lin, C.; Sun, Y. Effect of Dys-1 Mutation on Gene Expression Profile in Space-Flown Caenorhabditis elegans. Muscle Nerve 2018, 58, 114–122. [Google Scholar] [CrossRef]
- Zhao, L.; Gao, Y.; Mi, D.; Sun, Y. Mining Potential Biomarkers Associated with Space Flight in Caenorhabditis elegans Experienced Shenzhou-8 Mission with Multiple Feature Selection Techniques. Mutat. Res. 2016, 791–792, 27–34. [Google Scholar] [CrossRef]
- Kitano, H. Systems Biology: A Brief Overview. Science 2002, 295, 1662–1664. [Google Scholar] [CrossRef]
- Barabási, A.-L.; Oltvai, Z.N. Network Biology: Understanding the Cell’s Functional Organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef]
- Wang, C.; Sang, C.; Akira, H.; Noriaki, I.; Rong, L.; Yang, C.; Sun, Y.; Yi, Z.; Zhuang, F. Changes of Muscle-Related Genes and Proteins After Spaceflight in Caenorhabditis elegans. Prog. Biochem. Biophys. 2008, 35, 1195–1201. [Google Scholar]
- Kim, B.-S.; Alcantara, A.V., Jr.; Moon, J.-H.; Higashitani, A.; Higashitani, N.; Etheridge, T.; Szewczyk, N.J.; Deane, C.S.; Gaffney, C.J.; Higashibata, A.; et al. Comparative Analysis of Muscle Atrophy during Spaceflight, Nutritional Deficiency and Disuse in the Nematode Caenorhabditis elegans. Int. J. Mol. Sci. 2023, 24, 12640. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Lund, J.; Kiraly, M.; Duke, K.; Jiang, M.; Stuart, J.M.; Eizinger, A.; Wylie, B.N.; Davidson, G.S. A Gene Expression Map for Caenorhabditis elegans. Science 2001, 293, 2087–2092. [Google Scholar] [CrossRef] [PubMed]
- Owen, A.B.; Stuart, J.; Mach, K.; Villeneuve, A.M.; Kim, S. A Gene Recommender Algorithm to Identify Coexpressed Genes in C. elegans. Genome Res. 2003, 13, 1828–1837. [Google Scholar] [CrossRef]
- Narasimhan, K.; Lambert, S.A.; Yang, A.W.H.; Riddell, J.; Mnaimneh, S.; Zheng, H.; Albu, M.; Najafabadi, H.S.; Reece-Hoyes, J.S.; Fuxman Bass, J.I.; et al. Mapping and Analysis of Caenorhabditis elegans Transcription Factor Sequence Specificities. eLife 2015, 4, e06967. [Google Scholar] [CrossRef]
- Nanda, S.; Jacques, M.-A.; Wang, W.; Myers, C.L.; Yilmaz, L.S.; Walhout, A.J. Systems-Level Transcriptional Regulation of Caenorhabditis elegans Metabolism. Mol. Syst. Biol. 2023, 19, e11443. [Google Scholar] [CrossRef]
- Spencer, W.C.; Zeller, G.; Watson, J.D.; Henz, S.R.; Watkins, K.L.; McWhirter, R.D.; Petersen, S.; Sreedharan, V.T.; Widmer, C.; Jo, J.; et al. A Spatial and Temporal Map of C. elegans Gene Expression. Genome Res. 2011, 21, 325–341. [Google Scholar] [CrossRef]
- Pauli, F.; Liu, Y.; Kim, Y.A.; Chen, P.-J.; Kim, S.K. Chromosomal Clustering and GATA Transcriptional Regulation of Intestine-Expressed Genes in C. elegans. Development 2006, 133, 287–295. [Google Scholar] [CrossRef]
- Sun, X.; Chen, W.-D.; Wang, Y.-D. DAF-16/FOXO Transcription Factor in Aging and Longevity. Front. Pharmacol. 2017, 8, 548. [Google Scholar] [CrossRef]
- Willis, C.R.G.; Szewczyk, N.J.; Costes, S.V.; Udranszky, I.A.; Reinsch, S.S.; Etheridge, T.; Conley, C.A. Comparative Transcriptomics Identifies Neuronal and Metabolic Adaptations to Hypergravity and Microgravity in Caenorhabditis elegans. iScience 2020, 23, 101734. [Google Scholar] [CrossRef]
- Jiang, H.; Guo, R.; Powell-Coffman, J.A. The Caenorhabditis Elegans Hif-1 Gene Encodes a bHLH-PAS Protein That Is Required for Adaptation to Hypoxia. Proc. Natl. Acad. Sci. USA 2001, 98, 7916–7921. [Google Scholar] [CrossRef] [PubMed]
- Da Silveira, W.A.; Fazelinia, H.; Rosenthal, S.B.; Laiakis, E.C.; Kim, M.S.; Meydan, C.; Kidane, Y.; Rathi, K.S.; Smith, S.M.; Stear, B.; et al. Comprehensive Multi-Omics Analysis Reveals Mitochondrial Stress as a Central Biological Hub for Spaceflight Impact. Cell 2020, 183, 1185–1201.e20. [Google Scholar] [CrossRef] [PubMed]
- Tullet, J.M.A.; Green, J.W.; Au, C.; Benedetto, A.; Thompson, M.A.; Clark, E.; Gilliat, A.F.; Young, A.; Schmeisser, K.; Gems, D. The SKN-1/Nrf2 Transcription Factor Can Protect against Oxidative Stress and Increase Lifespan in C. elegans by Distinct Mechanisms. Aging Cell 2017, 16, 1191–1194. [Google Scholar] [CrossRef] [PubMed]
- Small, T.M.; Gernert, K.M.; Flaherty, D.B.; Mercer, K.B.; Borodovsky, M.; Benian, G.M. Three New Isoforms of Caenorhabditis elegans UNC-89 Containing MLCK-like Protein Kinase Domains. J. Mol. Biol. 2004, 342, 91–108. [Google Scholar] [CrossRef]
- Swoboda, P.; Adler, H.T.; Thomas, J.H. The RFX-Type Transcription Factor DAF-19 Regulates Sensory Neuron Cilium Formation in C. elegans. Mol. Cell 2000, 5, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Laranjeiro, R.; Harinath, G.; Pollard, A.K.; Gaffney, C.J.; Deane, C.S.; Vanapalli, S.A.; Etheridge, T.; Szewczyk, N.J.; Driscoll, M. Spaceflight Affects Neuronal Morphology and Alters Transcellular Degradation of Neuronal Debris in Adult Caenorhabditis elegans. iScience 2021, 24, 102105. [Google Scholar] [CrossRef] [PubMed]
- Higashitani, A.; Teranishi, M.; Nakagawa, Y.; Itoh, Y.; Sudevan, S.; Szewczyk, N.J.; Kubota, Y.; Abe, T.; Kobayashi, T. Increased Mitochondrial Ca2+ Contributes to Health Decline with Age and Duchene Muscular Dystrophy in C. elegans. FASEB J. 2023, 37, e22851. [Google Scholar] [CrossRef]
- Murgia, M.; Ciciliot, S.; Nagaraj, N.; Reggiani, C.; Schiaffino, S.; Franchi, M.V.; Pišot, R.; Šimunič, B.; Toniolo, L.; Blaauw, B.; et al. Signatures of Muscle Disuse in Spaceflight and Bed Rest Revealed by Single Muscle Fiber Proteomics. PNAS Nexus 2022, 1, gac086. [Google Scholar] [CrossRef]
- Deane, C.S.; Phillips, B.E.; Willis, C.R.G.; Wilkinson, D.J.; Smith, K.; Higashitani, N.; Williams, J.P.; Szewczyk, N.J.; Atherton, P.J.; Higashitani, A.; et al. Proteomic Features of Skeletal Muscle Adaptation to Resistance Exercise Training as a Function of Age. Geroscience 2023, 45, 1271–1287. [Google Scholar] [CrossRef]
- Ellwood, R.A.; Slade, L.; Lewis, J.; Torregrossa, R.; Sudevan, S.; Piasecki, M.; Whiteman, M.; Etheridge, T.; Szewczyk, N.J. Sulfur Amino Acid Supplementation Displays Therapeutic Potential in a C. elegans Model of Duchenne Muscular Dystrophy. Commun. Biol. 2022, 5, 1255. [Google Scholar] [CrossRef]
- Hrach, H.C.; O’Brien, S.; Steber, H.S.; Newbern, J.; Rawls, A.; Mangone, M. Transcriptome Changes during the Initiation and Progression of Duchenne Muscular Dystrophy in Caenorhabditis elegans. Hum. Mol. Genet. 2020, 29, 1607–1623. [Google Scholar] [CrossRef] [PubMed]
- Nakamori, M.; Takahashi, M.P. The Role of α-Dystrobrevin in Striated Muscle. Int. J. Mol. Sci. 2011, 12, 1660–1671. [Google Scholar] [CrossRef] [PubMed]
- Morange, M. A New Revolution? The Place of Systems Biology and Synthetic Biology in the History of Biology. EMBO Rep. 2009, 10 (Suppl. S1), S50–S53. [Google Scholar] [CrossRef]
- Shapira, P.; Kwon, S.; Youtie, J. Tracking the Emergence of Synthetic Biology. Scientometrics 2017, 112, 1439–1469. [Google Scholar] [CrossRef]
- Voigt, C.A. Synthetic Biology 2020-2030: Six Commercially-Available Products That Are Changing Our World. Nat. Commun. 2020, 11, 6379. [Google Scholar] [CrossRef]
- Sawyers, L.; Anderson, C.; Boyd, M.J.; Hessel, V.; Wotring, V.; Williams, P.M.; Toh, L.S. Astropharmacy: Pushing the Boundaries of the Pharmacists’ Role for Sustainable Space Exploration. Res. Soc. Adm. Pharm. 2022, 18, 3612–3621. [Google Scholar] [CrossRef]
- Aziz, S.; Raza, M.A.; Noreen, M.; Iqbal, M.Z.; Raza, S.M. Astropharmacy: Roles of Pharmacist in Space. Innov. Pharm. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Reichard, J.F.; Phelps, S.E.; Lehnhardt, K.R.; Young, M.; Easter, B.D. The Effect of Long-Term Spaceflight on Drug Potency and the Risk of Medication Failure. npj Microgravity 2023, 9, 35. [Google Scholar] [CrossRef]
- Mehta, P.; Bhayani, D. Impact of Space Environment on Stability of Medicines: Challenges and Prospects. J. Pharm. Biomed. Anal. 2017, 136, 111–119. [Google Scholar] [CrossRef]
- Blue, R.S.; Bayuse, T.M.; Daniels, V.R.; Wotring, V.E.; Suresh, R.; Mulcahy, R.A.; Antonsen, E.L. Supplying a Pharmacy for NASA Exploration Spaceflight: Challenges and Current Understanding. npj Microgravity 2019, 5, 14. [Google Scholar] [CrossRef]
- Wotring, V.E. Medication Use by U.S. Crewmembers on the International Space Station. FASEB J. 2015, 29, 4417–4423. [Google Scholar] [CrossRef] [PubMed]
- Santomartino, R.; Averesch, N.J.H.; Bhuiyan, M.; Cockell, C.S.; Colangelo, J.; Gumulya, Y.; Lehner, B.; Lopez-Ayala, I.; McMahon, S.; Mohanty, A.; et al. Toward Sustainable Space Exploration: A Roadmap for Harnessing the Power of Microorganisms. Nat. Commun. 2023, 14, 1391. [Google Scholar] [CrossRef] [PubMed]
- Ferranti, F.; Del Bianco, M.; Pacelli, C. Advantages and Limitations of Current Microgravity Platforms for Space Biology Research. NATO Adv. Sci. Inst. Ser. E Appl. Sci. 2020, 11, 68. [Google Scholar] [CrossRef]
- Stahl-Rommel, S.; Jain, M.; Nguyen, H.N.; Arnold, R.R.; Aunon-Chancellor, S.M.; Sharp, G.M.; Castro, C.L.; John, K.K.; Juul, S.; Turner, D.J.; et al. Real-Time Culture-Independent Microbial Profiling Onboard the International Space Station Using Nanopore Sequencing. Genes 2021, 12, 106. [Google Scholar] [CrossRef] [PubMed]
- Comfort, P.; McMahon, J.J.; Jones, P.A.; Cuthbert, M.; Kendall, K.; Lake, J.P.; Haff, G.G. Effects of Spaceflight on Musculoskeletal Health: A Systematic Review and Meta-Analysis, Considerations for Interplanetary Travel. Sports Med. 2021, 51, 2097–2114. [Google Scholar] [CrossRef]
- Scott, R.T.; Grigorev, K.; Mackintosh, G.; Gebre, S.G.; Mason, C.E.; Del Alto, M.E.; Costes, S.V. Advancing the Integration of Biosciences Data Sharing to Further Enable Space Exploration. Cell Rep. 2020, 33, 108441. [Google Scholar] [CrossRef]
- Ray, S.; Gebre, S.; Fogle, H.; Berrios, D.C.; Tran, P.B.; Galazka, J.M.; Costes, S.V. GeneLab: Omics Database for Spaceflight Experiments. Bioinformatics 2019, 35, 1753–1759. [Google Scholar] [CrossRef]
- Antunes, A.; Cockell, C.; De Saeger, S.; Grohman, E.; Kish, A.; Leys, N.; Moeller, R.; Moissl-Eichinger, C.; Nislow, C.; Pacelli, C.; et al. ROADMAP #9: Biology in Space and Analogue Environments; ESA: Paris, France, 2021; pp. 1–49. [Google Scholar]
- Quiroga Gutierrez, A.C.; Lindegger, D.J.; Taji Heravi, A.; Stojanov, T.; Sykora, M.; Elayan, S.; Mooney, S.J.; Naslund, J.A.; Fadda, M.; Gruebner, O. Reproducibility and Scientific Integrity of Big Data Research in Urban Public Health and Digital Epidemiology: A Call to Action. Int. J. Environ. Res. Public Health 2023, 20, 1473. [Google Scholar] [CrossRef]
- Arora, S.; Pattwell, S.S.; Holland, E.C.; Bolouri, H. Variability in Estimated Gene Expression among Commonly Used RNA-Seq Pipelines. Sci. Rep. 2020, 10, 2734. [Google Scholar] [CrossRef]
- Lee, P.H.U.; Chung, M.; Ren, Z.; Mair, D.B.; Kim, D.-H. Factors Mediating Spaceflight-Induced Skeletal Muscle Atrophy. Am. J. Physiol. Cell Physiol. 2022, 322, C567–C580. [Google Scholar] [CrossRef]
- Maffiuletti, N.A.; Green, D.A.; Vaz, M.A.; Dirks, M.L. Neuromuscular Electrical Stimulation as a Potential Countermeasure for Skeletal Muscle Atrophy and Weakness During Human Spaceflight. Front. Physiol. 2019, 10, 1031. [Google Scholar] [CrossRef] [PubMed]
- National Academies of Sciences, Engineering, and Medicine. Thriving in Space: Ensuring the Future of Biological and Physical Sciences Research: A Decadal Survey for 2023–2032; The National Academies Press: Washington, DA, USA, 2023; ISBN 9780309694988. [Google Scholar]
- Creech, S.; Guidi, J.; Elburn, D. Artemis: An Overview of NASA’s Activities to Return Humans to the Moon. In Proceedings of the 2022 IEEE Aerospace Conference (AERO), Big Sky, MT, USA, 5–12 March 2022; pp. 1–7. [Google Scholar]
- Higashitani, A.; Hashizume, T.; Sugimoto, T.; Mori, C.; Nemoto, K.; Etheridge, T.; Higashitani, N.; Takanami, T.; Suzuki, H.; Fukui, K.; et al. C. elegans RNAi Space Experiment (CERISE) in Japanese Experiment Module KIBO. Biol. Sci. Space 2009, 23, 183–187. [Google Scholar] [CrossRef] [PubMed]







| Strain | Ground | Flight | ||||||
|---|---|---|---|---|---|---|---|---|
| Sample Size | Diameter (µm) | Length (µm) | f95 (µN) | Sample Size | Diameter (µm) | Length (µm) | f95 (µN) | |
| wt | 30 | 46.97 ± 1.92 | 1077 ± 60 | 22.34 ± 5.67 | 30 | 44.35 ± 1.88 | 1062 ± 42 | 18.62 ± 4.05 |
| dys-1 | 25 | 48.24 ± 1.87 | 1110 ± 43 | 21.38 ± 5.39 | 29 | 44.41 ± 2.29 | 1080 ± 45 | 14.23 ± 3.87 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soni, P.; Edwards, H.; Anupom, T.; Rahman, M.; Lesanpezeshki, L.; Blawzdziewicz, J.; Cope, H.; Gharahdaghi, N.; Scott, D.; Toh, L.S.; et al. Spaceflight Induces Strength Decline in Caenorhabditis elegans. Cells 2023, 12, 2470. https://doi.org/10.3390/cells12202470
Soni P, Edwards H, Anupom T, Rahman M, Lesanpezeshki L, Blawzdziewicz J, Cope H, Gharahdaghi N, Scott D, Toh LS, et al. Spaceflight Induces Strength Decline in Caenorhabditis elegans. Cells. 2023; 12(20):2470. https://doi.org/10.3390/cells12202470
Chicago/Turabian StyleSoni, Purushottam, Hunter Edwards, Taslim Anupom, Mizanur Rahman, Leila Lesanpezeshki, Jerzy Blawzdziewicz, Henry Cope, Nima Gharahdaghi, Daniel Scott, Li Shean Toh, and et al. 2023. "Spaceflight Induces Strength Decline in Caenorhabditis elegans" Cells 12, no. 20: 2470. https://doi.org/10.3390/cells12202470