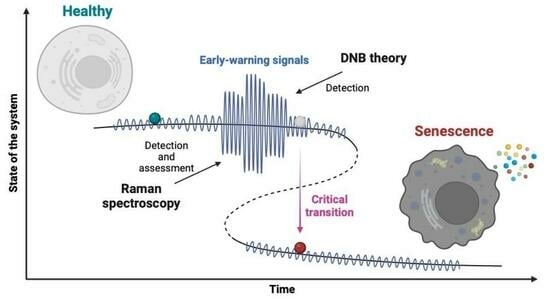
New Possibilities for Evaluating the Development of Age-Related Pathologies Using the Dynamical Network Biomarkers Theory
Abstract
:1. Introduction
2. Dynamical Network Biomarkers Theory
2.1. The Concept of DNB Theory
2.2. The Applications of DNB Theory
2.3. Cancer and Cellular Senescence
3. Raman Spectroscopy
3.1. A General Background on Raman Spectroscopy
3.2. Raman Spectroscopy and Cellular Senescence
3.3. Raman Imaging and DNB Analysis
4. Senolytics and Senomorphics
5. DNB Analysis in Metabolism
5.1. Identification of DNB Genes
5.2. Verification of DNB Genes Using a Drosophila Model
6. Homeostasis and Allostasis in Aging
7. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beard, J.R.; Si, Y.; Liu, Z.; Chenoweth, L.; Hanewald, K.; Lipsitz, L. Intrinsic Capacity: Validation of a New WHO Concept for Healthy Aging in a Longitudinal Chinese Study. J. Gerontol. Ser. A 2022, 77, 94–100. [Google Scholar] [CrossRef]
- Kirkland, J.L.; Stout, M.B.; Sierra, F. Resilience in Aging Mice. J. Gerontol. A Biol. Sci. Med. Sci. 2016, 71, 1407–1414. [Google Scholar] [CrossRef]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of aging: An expanding universe. Cell 2022, 186, 243–278. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, B.K.; Berger, S.L.; Brunet, A.; Campisi, J.; Cuervo, A.M.; Epel, E.S.; Franceschi, C.; Lithgow, G.J.; Morimoto, R.I.; Pessin, J.E.; et al. Geroscience: Linking aging to chronic disease. Cell 2014, 159, 709–713. [Google Scholar] [CrossRef] [PubMed]
- Baechle, J.J.; Chen, N.; Makhijani, P.; Winer, S.; Furman, D.; Winer, D.A. Chronic inflammation and the hallmarks of aging. Mol. Metab. 2023, 74, 101755. [Google Scholar] [CrossRef] [PubMed]
- Gorgoulis, V.; Adams, P.D.; Alimonti, A.; Bennett, D.C.; Bischof, O.; Bishop, C.; Campisi, J.; Collado, M.; Evangelou, K.; Ferbeyre, G.; et al. Cellular Senescence: Defining a Path Forward. Cell 2019, 179, 813–827. [Google Scholar] [CrossRef] [PubMed]
- Roninson, I.B.; Broude, E.V.; Chang, B.D. If not apoptosis, then what? Treatment-induced senescence and mitotic catastrophe in tumor cells. Drug Resist. Updat. 2001, 4, 303–313. [Google Scholar] [CrossRef]
- Hernandez-Segura, A.; Nehme, J.; Demaria, M. Hallmarks of Cellular Senescence. Trends Cell Biol. 2018, 28, 436–453. [Google Scholar] [CrossRef]
- Gurkar, A.U.; Gerencser, A.A.; Mora, A.L.; Nelson, A.C.; Zhang, A.R.; Lagnado, A.B.; Enninful, A.; Benz, C.; Furman, D.; Beaulieu, D.; et al. Spatial mapping of cellular senescence: Emerging challenges and opportunities. Nat. Aging 2023, 3, 776–790. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, R.; Liu, Z.P.; Li, M.; Aihara, K. Detecting early-warning signals for sudden deterioration of complex diseases by dynamical network biomarkers. Sci. Rep. 2012, 2, 342. [Google Scholar] [CrossRef]
- Sterling, P. Allostasis: A New Paradigm to Explain Arousal Pathology. In Handbook of Life Stress, Cognition and Health; Wiley: Hoboken, NJ, USA, 1988. [Google Scholar]
- Scheffer, M.; Bascompte, J.; Brock, W.A.; Brovkin, V.; Carpenter, S.R.; Dakos, V.; Held, H.; van Nes, E.H.; Rietkerk, M.; Sugihara, G. Early-warning signals for critical transitions. Nature 2009, 461, 53–59. [Google Scholar] [CrossRef]
- Scheffer, M.; Carpenter, S.R.; Lenton, T.M.; Bascompte, J.; Brock, W.; Dakos, V.; van de Koppel, J.; van de Leemput, I.A.; Levin, S.A.; van Nes, E.H.; et al. Anticipating critical transitions. Science 2012, 338, 344–348. [Google Scholar] [CrossRef]
- Tytgat, H.L.P.; Nobrega, F.L.; van der Oost, J.; de Vos, W.M. Bowel Biofilms: Tipping Points between a Healthy and Compromised Gut? Trends Microbiol. 2019, 27, 17–25. [Google Scholar] [CrossRef]
- Carpenter, S.R.; Brock, W.A. Rising variance: A leading indicator of ecological transition. Ecol. Lett. 2006, 9, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Dakos, V.; Scheffer, M.; van Nes, E.H.; Brovkin, V.; Petoukhov, V.; Held, H. Slowing down as an early warning signal for abrupt climate change. Proc. Natl. Acad. Sci. USA 2008, 105, 14308–14312. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.; Lu, T.C. Network catastrophe: Self-organized patterns reveal both the instability and the structure of complex networks. Sci. Rep. 2015, 5, 9450. [Google Scholar] [CrossRef] [PubMed]
- Veraart, A.J.; Faassen, E.J.; Dakos, V.; van Nes, E.H.; Lurling, M.; Scheffer, M. Recovery rates reflect distance to a tipping point in a living system. Nature 2011, 481, 357–359. [Google Scholar] [CrossRef]
- Wissel, C. A universal law of the characteristic return time near thresholds. Oecologia 1984, 65, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Tripati, A.; Backman, J.; Elderfield, H.; Ferretti, P. Eocene bipolar glaciation associated with global carbon cycle changes. Nature 2005, 436, 341–346. [Google Scholar] [CrossRef]
- Aihara, K.; Liu, R.; Koizumi, K.; Liu, X.; Chen, L. Dynamical network biomarkers: Theory and applications. Gene 2022, 808, 145997. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; van Oudenaarden, A. Nature, nurture, or chance: Stochastic gene expression and its consequences. Cell 2008, 135, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chang, X.; Leng, S.; Tang, H.; Aihara, K.; Chen, L. Detection for disease tipping points by landscape dynamic network biomarkers. Natl. Sci. Rev. 2019, 6, 775–785. [Google Scholar] [CrossRef]
- Liu, R.; Zhong, J.; Hong, R.; Chen, E.; Aihara, K.; Chen, P.; Chen, L. Predicting local COVID-19 outbreaks and infectious disease epidemics based on landscape network entropy. Sci. Bull. 2021, 66, 2265–2270. [Google Scholar] [CrossRef]
- Gao, R.; Yan, J.; Li, P.; Chen, L. Detecting the critical states during disease development based on temporal network flow entropy. Brief. Bioinform. 2022, 23, bbac164. [Google Scholar] [CrossRef]
- Yang, B.; Li, M.; Tang, W.; Liu, W.; Zhang, S.; Chen, L.; Xia, J. Dynamic network biomarker indicates pulmonary metastasis at the tipping point of hepatocellular carcinoma. Nat. Commun. 2018, 9, 678. [Google Scholar] [CrossRef]
- Liu, R.; Wang, J.; Ukai, M.; Sewon, K.; Chen, P.; Suzuki, Y.; Wang, H.; Aihara, K.; Okada-Hatakeyama, M.; Chen, L. Hunt for the tipping point during endocrine resistance process in breast cancer by dynamic network biomarkers. J. Mol. Cell Biol. 2019, 11, 649–664. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, H.; Ge, J.; Mi, T.; Cui, X.; Tu, F.; Gu, X.; Zeng, T.; Chen, L. Landscape dynamic network biomarker analysis reveals the tipping point of transcriptome reprogramming to prevent skin photodamage. J. Mol. Cell Biol. 2022, 13, 822–833. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Han, X.; Chen, Y.; Tong, X.; Xue, Y.; Yao, S.; Tang, S.; Pan, Y.; Sun, Y.; Wang, X.; et al. Oxidative stress-triggered Wnt signaling perturbation characterizes the tipping point of lung adeno-to-squamous transdifferentiation. Signal Transduct. Target. Ther. 2023, 8, 16. [Google Scholar] [CrossRef]
- Freedman, S.L.; Xu, B.; Goyal, S.; Mani, M. A dynamical systems treatment of transcriptomic trajectories in hematopoiesis. Development 2023, 150, dev201280. [Google Scholar] [CrossRef]
- Li, L.; Xu, Y.; Yan, L.; Li, X.; Li, F.; Liu, Z.; Zhang, C.; Lou, Y.; Gao, D.; Cheng, X.; et al. Dynamic network biomarker factors orchestrate cell-fate determination at tipping points during hESC differentiation. Innovation 2023, 4, 100364. [Google Scholar] [CrossRef] [PubMed]
- Haruki, T.; Yonezawa, S.; Koizumi, K.; Yoshida, Y.; Watanabe, T.M.; Fujita, H.; Oshima, Y.; Oku, M.; Taketani, A.; Yamazaki, M.; et al. Application of the Dynamical Network Biomarker Theory to Raman Spectra. Biomolecules 2022, 12, 1730. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, K.; Oku, M.; Hayashi, S.; Inujima, A.; Shibahara, N.; Chen, L.; Igarashi, Y.; Tobe, K.; Saito, S.; Kadowaki, M.; et al. Identifying pre-disease signals before metabolic syndrome in mice by dynamical network biomarkers. Sci. Rep. 2019, 9, 8767. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, K.; Oku, M.; Hayashi, S.; Inujima, A.; Shibahara, N.; Chen, L.; Igarashi, Y.; Tobe, K.; Saito, S.; Kadowaki, M.; et al. Suppression of Dynamical Network Biomarker Signals at the Predisease State (Mibyou) before Metabolic Syndrome in Mice by a Traditional Japanese Medicine (Kampo Formula) Bofutsushosan. Evid. Based Complement. Altern. Med. 2020, 2020, 9129134. [Google Scholar] [CrossRef]
- Yang, Y.; Tian, Z.; Song, M.; Ma, C.; Ge, Z.; Li, P. Detecting the Critical States of Type 2 Diabetes Mellitus Based on Degree Matrix Network Entropy by Cross-Tissue Analysis. Entropy 2022, 24, 1249. [Google Scholar] [CrossRef]
- Rukhlenko, O.S.; Halasz, M.; Rauch, N.; Zhernovkov, V.; Prince, T.; Wynne, K.; Maher, S.; Kashdan, E.; MacLeod, K.; Carragher, N.O.; et al. Control of cell state transitions. Nature 2022, 609, 975–985. [Google Scholar] [CrossRef]
- Erenpreisa, J.; Salmina, K.; Cragg, M.S. Accelerated Senescence of Cancer Stem Cells: A Failure to Thrive or a Route to Survival? In Senescence; Jolanta, D., Wojciech, K., Eds.; IntechOpen: Rijeka, Croatia, 2017; p. Ch.4. [Google Scholar]
- Serrano, M.; Lin, A.W.; McCurrach, M.E.; Beach, D.; Lowe, S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997, 88, 593–602. [Google Scholar] [CrossRef]
- Schmitt, C.A.; Fridman, J.S.; Yang, M.; Lee, S.; Baranov, E.; Hoffman, R.M.; Lowe, S.W. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell 2002, 109, 335–346. [Google Scholar] [CrossRef]
- Saleh, T.; Bloukh, S.; Carpenter, V.J.; Alwohoush, E.; Bakeer, J.; Darwish, S.; Azab, B.; Gewirtz, D.A. Therapy-Induced Senescence: An “Old” Friend Becomes the Enemy. Cancers 2020, 12, 822. [Google Scholar] [CrossRef]
- Milanovic, M.; Fan, D.N.Y.; Belenki, D.; Dabritz, J.H.M.; Zhao, Z.; Yu, Y.; Dorr, J.R.; Dimitrova, L.; Lenze, D.; Monteiro Barbosa, I.A.; et al. Senescence-associated reprogramming promotes cancer stemness. Nature 2018, 553, 96–100. [Google Scholar] [CrossRef]
- Lee, S.; Schmitt, C.A. The dynamic nature of senescence in cancer. Nat. Cell Biol. 2019, 21, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Erenpreisa, J.; Salmina, K.; Anatskaya, O.; Cragg, M.S. Paradoxes of cancer: Survival at the brink. Semin. Cancer Biol. 2022, 81, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Jackson, T.R.; Salmina, K.; Huna, A.; Inashkina, I.; Jankevics, E.; Riekstina, U.; Kalnina, Z.; Ivanov, A.; Townsend, P.A.; Cragg, M.S.; et al. DNA damage causes TP53-dependent coupling of self-renewal and senescence pathways in embryonal carcinoma cells. Cell Cycle 2013, 12, 430–441. [Google Scholar] [CrossRef]
- Huna, A.; Salmina, K.; Erenpreisa, J.; Vazquez-Martin, A.; Krigerts, J.; Inashkina, I.; Gerashchenko, B.I.; Townsend, P.A.; Cragg, M.S.; Jackson, T.R. Role of stress-activated OCT4A in the cell fate decisions of embryonal carcinoma cells treated with etoposide. Cell Cycle 2015, 14, 2969–2984. [Google Scholar] [CrossRef]
- Kou, F.; Wu, L.; Ren, X.; Yang, L. Chromosome Abnormalities: New Insights into Their Clinical Significance in Cancer. Mol. Ther. Oncolytics 2020, 17, 562–570. [Google Scholar] [CrossRef]
- Was, H.; Borkowska, A.; Olszewska, A.; Klemba, A.; Marciniak, M.; Synowiec, A.; Kieda, C. Polyploidy formation in cancer cells: How a Trojan horse is born. Semin. Cancer Biol. 2022, 81, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Erenpreisa, J.; Sikora, E. Polyploid giant cancer cells: An emerging new field of cancer biology. Semin. Cancer Biol. 2022, 81, 1–4. [Google Scholar] [CrossRef]
- Liu, J. The dualistic origin of human tumors. Semin. Cancer Biol. 2018, 53, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Huna, A.; Salmina, K.; Jascenko, E.; Duburs, G.; Inashkina, I.; Erenpreisa, J. Self-Renewal Signalling in Presenescent Tetraploid IMR90 Cells. J. Aging Res. 2011, 2011, 103253. [Google Scholar] [CrossRef] [PubMed]
- Kocarnik, J.M.; Compton, K.; Dean, F.E.; Fu, W.; Gaw, B.L.; Harvey, J.D.; Henrikson, H.J.; Lu, D.; Pennini, A.; Xu, R.; et al. Cancer Incidence, Mortality, Years of Life Lost, Years Lived with Disability, and Disability-Adjusted Life Years for 29 Cancer Groups From 2010 to 2019: A Systematic Analysis for the Global Burden of Disease Study 2019. JAMA Oncol. 2022, 8, 420–444. [Google Scholar] [CrossRef]
- Liendl, L.; Grillari, J.; Schosserer, M. Raman fingerprints as promising markers of cellular senescence and aging. Geroscience 2020, 42, 377–387. [Google Scholar] [CrossRef]
- Liu, K.; Zhao, Q.; Li, B.; Zhao, X. Raman Spectroscopy: A Novel Technology for Gastric Cancer Diagnosis. Front. Bioeng. Biotechnol. 2022, 10, 856591. [Google Scholar] [CrossRef] [PubMed]
- Okada, M.; Smith, N.I.; Palonpon, A.F.; Endo, H.; Kawata, S.; Sodeoka, M.; Fujita, K. Label-free Raman observation of cytochrome c dynamics during apoptosis. Proc. Natl. Acad. Sci. USA 2012, 109, 28–32. [Google Scholar] [CrossRef]
- Ichimura, T.; Chiu, L.-d.; Fujita, K.; Kawata, S.; Watanabe, T.M.; Yanagida, T.; Fujita, H. Visualizing Cell State Transition Using Raman Spectroscopy. PLoS ONE 2014, 9, e84478. [Google Scholar] [CrossRef]
- Ichimura, T.; Chiu, L.-d.; Fujita, K.; Machiyama, H.; Kawata, S.; Watanabe, T.M.; Fujita, H. Visualizing the appearance and disappearance of the attractor of differentiation using Raman spectral imaging. Sci. Rep. 2015, 5, 11358. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, T.; Chiu, L.-d.; Fujita, K.; Machiyama, H.; Yamaguchi, T.; Watanabe, T.M.; Fujita, H. Non-label immune cell state prediction using Raman spectroscopy. Sci. Rep. 2016, 6, 37562. [Google Scholar] [CrossRef]
- Chaudhary, N.; Nguyen, T.N.Q.; Cullen, D.; Meade, A.D.; Wynne, C. Discrimination of immune cell activation using Raman micro-spectroscopy in an in-vitro & ex-vivo model. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 248, 119118. [Google Scholar] [CrossRef]
- Taketani, A.; Andriana, B.B.; Matsuyoshi, H.; Sato, H. Raman endoscopy for monitoring the anticancer drug treatment of colorectal tumors in live mice. Analyst 2017, 142, 3680–3688. [Google Scholar] [CrossRef] [PubMed]
- Guerrini, L.; Garcia-Rico, E.; O’Loghlen, A.; Giannini, V.; Alvarez-Puebla, R.A. Surface-Enhanced Raman Scattering (SERS) Spectroscopy for Sensing and Characterization of Exosomes in Cancer Diagnosis. Cancers 2021, 13, 2179. [Google Scholar] [CrossRef]
- Ogawa, K.; Oshima, Y.; Etoh, T.; Kaisyakuji, Y.; Tojigamori, M.; Ohno, Y.; Shiraishi, N.; Inomata, M. Label-free detection of human enteric nerve system using Raman spectroscopy: A pilot study for diagnosis of Hirschsprung disease. J. Pediatr. Surg. 2021, 56, 1150–1156. [Google Scholar] [CrossRef] [PubMed]
- Gamsjaeger, S.; Eriksen, E.F.; Paschalis, E.P. Effect of hormone replacement therapy on bone formation quality and mineralization regulation mechanisms in early postmenopausal women. Bone Rep. 2021, 14, 101055. [Google Scholar] [CrossRef] [PubMed]
- Asaoka, R.; Kiyomatsu, H.; Miura, H.; Jono, A.; Kinoshita, T.; Takao, M.; Katagiri, T.; Oshima, Y. Prognostic potential and pathological validation of a diagnostic application using Raman spectroscopy in the characterization of degenerative changes in the cartilage of the humeral head. J. Biomed. Opt. 2022, 27, 115002. [Google Scholar] [CrossRef]
- Oshima, Y.; Haruki, T.; Koizumi, K.; Yonezawa, S.; Taketani, A.; Kadowaki, M.; Saito, S. Practices, Potential, and Perspectives for Detecting Predisease Using Raman Spectroscopy. Int. J. Mol. Sci. 2023, 24, 12170. [Google Scholar] [CrossRef]
- Liendl, L.; Schosserer, M. Raman microspectroscopy: Sub-cellular chemical imaging of aging. Aging 2021, 13, 24922–24923. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Li, H.; Han, Z.; Zhang, C.; Zhao, J.; Miao, C.; Yan, S.; Mao, A.; Zhao, H.; Han, Z. Label-free assessment of replicative senescence in mesenchymal stem cells by Raman microspectroscopy. Biomed. Opt. Express 2015, 6, 4493–4500. [Google Scholar] [CrossRef]
- Eberhardt, K.; Beleites, C.; Marthandan, S.; Matthaus, C.; Diekmann, S.; Popp, J. Raman and Infrared Spectroscopy Distinguishing Replicative Senescent from Proliferating Primary Human Fibroblast Cells by Detecting Spectral Differences Mainly Due to Biomolecular Alterations. Anal. Chem. 2017, 89, 2937–2947. [Google Scholar] [CrossRef]
- Eberhardt, K.; Matthäus, C.; Winter, D.; Wiegand, C.; Hipler, U.-C.; Diekmann, S.; Popp, J. Raman and infrared spectroscopy differentiate senescent from proliferating cells in a human dermal fibroblast 3D skin model. Analyst 2017, 142, 4405–4414. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Lee, C.; Yang, W.; Li, A.; Mukherjee, A.; Basan, M.; Ran, C.; Yin, W.; Tabin, C.J.; Fu, D.; et al. Protein and lipid mass concentration measurement in tissues by stimulated Raman scattering microscopy. Proc. Natl. Acad. Sci. USA 2022, 119, e2117938119. [Google Scholar] [CrossRef]
- Liu, X.; Oh, S.; Kirschner, M.W. The uniformity and stability of cellular mass density in mammalian cell culture. Front. Cell Dev. Biol. 2022, 10, 1017499. [Google Scholar] [CrossRef]
- Neurohr, G.E.; Terry, R.L.; Lengefeld, J.; Bonney, M.; Brittingham, G.P.; Moretto, F.; Miettinen, T.P.; Vaites, L.P.; Soares, L.M.; Paulo, J.A.; et al. Excessive Cell Growth Causes Cytoplasm Dilution and Contributes to Senescence. Cell 2019, 176, 1083–1097.e1018. [Google Scholar] [CrossRef]
- Zhang, L.; Pitcher, L.E.; Prahalad, V.; Niedernhofer, L.J.; Robbins, P.D. Targeting cellular senescence with senotherapeutics: Senolytics and senomorphics. FEBS J. 2023, 290, 1362–1383. [Google Scholar] [CrossRef]
- Ravichandra, A.; Filliol, A.; Schwabe, R.F. Chimeric Antigen Receptor T Cells as Senolytic and Antifibrotic Therapy. Hepatology 2021, 73, 1227–1229. [Google Scholar] [CrossRef] [PubMed]
- Grosse, L.; Wagner, N.; Emelyanov, A.; Molina, C.; Lacas-Gervais, S.; Wagner, K.D.; Bulavin, D.V. Defined p16(High) Senescent Cell Types Are Indispensable for Mouse Healthspan. Cell Metab. 2020, 32, 87–99.e86. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Tchkonia, T.; Pirtskhalava, T.; Gower, A.C.; Ding, H.; Giorgadze, N.; Palmer, A.K.; Ikeno, Y.; Hubbard, G.B.; Lenburg, M.; et al. The Achilles’ heel of senescent cells: From transcriptome to senolytic drugs. Aging Cell 2015, 14, 644–658. [Google Scholar] [CrossRef]
- Zhu, Y.; Doornebal, E.J.; Pirtskhalava, T.; Giorgadze, N.; Wentworth, M.; Fuhrmann-Stroissnigg, H.; Niedernhofer, L.J.; Robbins, P.D.; Tchkonia, T.; Kirkland, J.L. New agents that target senescent cells: The flavone, fisetin, and the BCL-X(L) inhibitors, A1331852 and A1155463. Aging 2017, 9, 955–963. [Google Scholar] [CrossRef]
- Zhu, Y.; Tchkonia, T.; Fuhrmann-Stroissnigg, H.; Dai, H.M.; Ling, Y.Y.; Stout, M.B.; Pirtskhalava, T.; Giorgadze, N.; Johnson, K.O.; Giles, C.B.; et al. Identification of a novel senolytic agent, navitoclax, targeting the Bcl-2 family of anti-apoptotic factors. Aging Cell 2016, 15, 428–435. [Google Scholar] [CrossRef]
- Xu, Q.; Fu, Q.; Li, Z.; Liu, H.; Wang, Y.; Lin, X.; He, R.; Zhang, X.; Ju, Z.; Campisi, J.; et al. The flavonoid procyanidin C1 has senotherapeutic activity and increases lifespan in mice. Nat. Metab. 2021, 3, 1706–1726. [Google Scholar] [CrossRef] [PubMed]
- Fuhrmann-Stroissnigg, H.; Ling, Y.Y.; Zhao, J.; McGowan, S.J.; Zhu, Y.; Brooks, R.W.; Grassi, D.; Gregg, S.Q.; Stripay, J.L.; Dorronsoro, A.; et al. Identification of HSP90 inhibitors as a novel class of senolytics. Nat. Commun. 2017, 8, 422. [Google Scholar] [CrossRef]
- Chaib, S.; Tchkonia, T.; Kirkland, J.L. Cellular senescence and senolytics: The path to the clinic. Nat. Med. 2022, 28, 1556–1568. [Google Scholar] [CrossRef]
- Baar, M.P.; Brandt, R.M.C.; Putavet, D.A.; Klein, J.D.D.; Derks, K.W.J.; Bourgeois, B.R.M.; Stryeck, S.; Rijksen, Y.; van Willigenburg, H.; Feijtel, D.A.; et al. Targeted Apoptosis of Senescent Cells Restores Tissue Homeostasis in Response to Chemotoxicity and Aging. Cell 2017, 169, 132–147.e116. [Google Scholar] [CrossRef]
- Guerrero, A.; Herranz, N.; Sun, B.; Wagner, V.; Gallage, S.; Guiho, R.; Wolter, K.; Pombo, J.; Irvine, E.E.; Innes, A.J.; et al. Cardiac glycosides are broad-spectrum senolytics. Nat. Metab. 2019, 1, 1074–1088. [Google Scholar] [CrossRef] [PubMed]
- Triana-Martínez, F.; Picallos-Rabina, P.; Da Silva-Álvarez, S.; Pietrocola, F.; Llanos, S.; Rodilla, V.; Soprano, E.; Pedrosa, P.; Ferreirós, A.; Barradas, M.; et al. Identification and characterization of Cardiac Glycosides as senolytic compounds. Nat. Commun. 2019, 10, 4731. [Google Scholar] [CrossRef] [PubMed]
- Johmura, Y.; Yamanaka, T.; Omori, S.; Wang, T.W.; Sugiura, Y.; Matsumoto, M.; Suzuki, N.; Kumamoto, S.; Yamaguchi, K.; Hatakeyama, S.; et al. Senolysis by glutaminolysis inhibition ameliorates various age-associated disorders. Science 2021, 371, 265–270. [Google Scholar] [CrossRef]
- Farr, J.N.; Xu, M.; Weivoda, M.M.; Monroe, D.G.; Fraser, D.G.; Onken, J.L.; Negley, B.A.; Sfeir, J.G.; Ogrodnik, M.B.; Hachfeld, C.M.; et al. Targeting cellular senescence prevents age-related bone loss in mice. Nat. Med. 2017, 23, 1072–1079. [Google Scholar] [CrossRef]
- Weichhart, T. mTOR as Regulator of Lifespan, Aging, and Cellular Senescence: A Mini-Review. Gerontology 2018, 64, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Yu, Z.; Sunchu, B.; Shoaf, J.; Dang, I.; Zhao, S.; Caples, K.; Bradley, L.; Beaver, L.M.; Ho, E.; et al. Rapamycin inhibits the secretory phenotype of senescent cells by a Nrf2-independent mechanism. Aging Cell 2017, 16, 564–574. [Google Scholar] [CrossRef]
- Moiseeva, O.; Deschênes-Simard, X.; St-Germain, E.; Igelmann, S.; Huot, G.; Cadar, A.E.; Bourdeau, V.; Pollak, M.N.; Ferbeyre, G. Metformin inhibits the senescence-associated secretory phenotype by interfering with IKK/NF-κB activation. Aging Cell 2013, 12, 489–498. [Google Scholar] [CrossRef]
- Noren Hooten, N.; Martin-Montalvo, A.; Dluzen, D.F.; Zhang, Y.; Bernier, M.; Zonderman, A.B.; Becker, K.G.; Gorospe, M.; de Cabo, R.; Evans, M.K. Metformin-mediated increase in DICER1 regulates microRNA expression and cellular senescence. Aging Cell 2016, 15, 572–581. [Google Scholar] [CrossRef]
- Jiang, X.; Ruan, X.L.; Xue, Y.X.; Yang, S.; Shi, M.; Wang, L.N. Metformin Reduces the Senescence of Renal Tubular Epithelial Cells in Diabetic Nephropathy via the MBNL1/miR-130a-3p/STAT3 Pathway. Oxid. Med. Cell Longev. 2020, 2020, 8708236. [Google Scholar] [CrossRef]
- Ali, D.; Chen, L.; Kowal, J.M.; Okla, M.; Manikandan, M.; AlShehri, M.; AlMana, Y.; AlObaidan, R.; AlOtaibi, N.; Hamam, R.; et al. Resveratrol inhibits adipocyte differentiation and cellular senescence of human bone marrow stromal stem cells. Bone 2020, 133, 115252. [Google Scholar] [CrossRef]
- Csiszar, A.; Sosnowska, D.; Wang, M.; Lakatta, E.G.; Sonntag, W.E.; Ungvari, Z. Age-associated proinflammatory secretory phenotype in vascular smooth muscle cells from the non-human primate Macaca mulatta: Reversal by resveratrol treatment. J. Gerontol. A Biol. Sci. Med. Sci. 2012, 67, 811–820. [Google Scholar] [CrossRef]
- Hubbard, B.P.; Sinclair, D.A. Small molecule SIRT1 activators for the treatment of aging and age-related diseases. Trends Pharmacol. Sci. 2014, 35, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.J.; Martin-Montalvo, A.; Mercken, E.M.; Palacios, H.H.; Ward, T.M.; Abulwerdi, G.; Minor, R.K.; Vlasuk, G.P.; Ellis, J.L.; Sinclair, D.A.; et al. The SIRT1 activator SRT1720 extends lifespan and improves health of mice fed a standard diet. Cell Rep. 2014, 6, 836–843. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, J.; Mu, X.; McGowan, S.J.; Angelini, L.; O’Kelly, R.D.; Yousefzadeh, M.J.; Sakamoto, A.; Aversa, Z.; LeBrasseur, N.K.; et al. Novel small molecule inhibition of IKK/NF-κB activation reduces markers of senescence and improves healthspan in mouse models of aging. Aging Cell 2021, 20, e13486. [Google Scholar] [CrossRef] [PubMed]
- Freund, A.; Patil, C.K.; Campisi, J. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J. 2011, 30, 1536–1548. [Google Scholar] [CrossRef]
- Alimbetov, D.; Davis, T.; Brook, A.J.; Cox, L.S.; Faragher, R.G.; Nurgozhin, T.; Zhumadilov, Z.; Kipling, D. Suppression of the senescence-associated secretory phenotype (SASP) in human fibroblasts using small molecule inhibitors of p38 MAP kinase and MK2. Biogerontology 2016, 17, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Griveau, A.; Wiel, C.; Ziegler, D.V.; Bergo, M.O.; Bernard, D. The JAK1/2 inhibitor ruxolitinib delays premature aging phenotypes. Aging Cell 2020, 19, e13122. [Google Scholar] [CrossRef]
- Wu, W.; Fu, J.; Gu, Y.; Wei, Y.; Ma, P.; Wu, J. JAK2/STAT3 regulates estrogen-related senescence of bone marrow stem cells. J. Endocrinol. 2020, 245, 141–153. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, L.; Lu, A.; Han, Y.; Colangelo, D.; Bukata, C.; Scibetta, A.; Yousefzadeh, M.J.; Li, X.; Gurkar, A.U.; et al. ATM is a key driver of NF-κB-dependent DNA-damage-induced senescence, stem cell dysfunction and aging. Aging 2020, 12, 4688–4710. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.T.; Park, J.T.; Choi, K.; Kim, Y.; Choi, H.J.C.; Jung, C.W.; Lee, Y.S.; Park, S.C. Chemical screening identifies ATM as a target for alleviating senescence. Nat. Chem. Biol. 2017, 13, 616–623. [Google Scholar] [CrossRef]
- Ota, H.; Eto, M.; Kano, M.R.; Kahyo, T.; Setou, M.; Ogawa, S.; Iijima, K.; Akishita, M.; Ouchi, Y. Induction of endothelial nitric oxide synthase, SIRT1, and catalase by statins inhibits endothelial senescence through the Akt pathway. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 2205–2211. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Uppal, H.; Demaria, M.; Desprez, P.Y.; Campisi, J.; Kapahi, P. Simvastatin suppresses breast cancer cell proliferation induced by senescent cells. Sci. Rep. 2015, 5, 17895. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.; Park, H.; Kim, H.P. Effects of flavonoids on senescence-associated secretory phenotype formation from bleomycin-induced senescence in BJ fibroblasts. Biochem. Pharmacol. 2015, 96, 337–348. [Google Scholar] [CrossRef]
- Paramos-de-Carvalho, D.; Jacinto, A.; Saúde, L. The right time for senescence. eLife 2021, 10, e72449. [Google Scholar] [CrossRef] [PubMed]
- Demaria, M.; Ohtani, N.; Youssef, S.A.; Rodier, F.; Toussaint, W.; Mitchell, J.R.; Laberge, R.M.; Vijg, J.; Van Steeg, H.; Dollé, M.E.; et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev. Cell 2014, 31, 722–733. [Google Scholar] [CrossRef]
- Suzuki, W.; Iizuka, S.; Tabuchi, M.; Funo, S.; Yanagisawa, T.; Kimura, M.; Sato, T.; Endo, T.; Kawamura, H. A new mouse model of spontaneous diabetes derived from ddY strain. Exp. Anim. 1999, 48, 181–189. [Google Scholar] [CrossRef]
- Zitzmann, M. Testosterone deficiency, insulin resistance and the metabolic syndrome. Nat. Rev. Endocrinol. 2009, 5, 673–681. [Google Scholar] [CrossRef]
- Emont, M.P.; Jacobs, C.; Essene, A.L.; Pant, D.; Tenen, D.; Colleluori, G.; Di Vincenzo, A.; Jorgensen, A.M.; Dashti, H.; Stefek, A.; et al. A single-cell atlas of human and mouse white adipose tissue. Nature 2022, 603, 926–933. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Yamamoto, S. Role of Drosophila in Human Disease Research 2.0. Int. J. Mol. Sci. 2022, 23, 4203. [Google Scholar] [CrossRef]
- Wangler, M.F.; Yamamoto, S.; Bellen, H.J. Fruit flies in biomedical research. Genetics 2015, 199, 639–653. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, H.; Du, J.; Tai, H.; Han, X.; Huang, N.; Wang, X.; Gong, H.; Yang, M.; Xiao, H. Glutamine Availability Regulates the Development of Aging Mediated by mTOR Signaling and Autophagy. Front. Pharmacol. 2022, 13, 924081. [Google Scholar] [CrossRef]
- Ito, T.; Igaki, T. Dissecting cellular senescence and SASP in Drosophila. Inflamm. Regen. 2016, 36, 25. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Ohsawa, S.; Igaki, T. Mitochondrial defects trigger proliferation of neighbouring cells via a senescence-associated secretory phenotype in Drosophila. Nat. Commun. 2014, 5, 5264. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Partridge, L. Promoting health and longevity through diet: From model organisms to humans. Cell 2015, 161, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Perrimon, N. What fuels the fly: Energy metabolism in Drosophila and its application to the study of obesity and diabetes. Sci. Adv. 2021, 7, eabg4336. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.A.; Chamoli, M.; Hilsabeck, T.A.; Pandey, M.; Bansal, S.; Chawla, G.; Kapahi, P. Evaluating the beneficial effects of dietary restrictions: A framework for precision nutrigeroscience. Cell Metab. 2021, 33, 2142–2173. [Google Scholar] [CrossRef]
- Oka, M.; Suzuki, E.; Asada, A.; Saito, T.; Iijima, K.M.; Ando, K. Increasing neuronal glucose uptake attenuates brain aging and promotes life span under dietary restriction in Drosophila. iScience 2021, 24, 101979. [Google Scholar] [CrossRef]
- Katewa, S.D.; Demontis, F.; Kolipinski, M.; Hubbard, A.; Gill, M.S.; Perrimon, N.; Melov, S.; Kapahi, P. Intramyocellular fatty-acid metabolism plays a critical role in mediating responses to dietary restriction in Drosophila melanogaster. Cell Metab. 2012, 16, 97–103. [Google Scholar] [CrossRef]
- Akagi, K.; Wilson, K.A.; Katewa, S.D.; Ortega, M.; Simons, J.; Hilsabeck, T.A.; Kapuria, S.; Sharma, A.; Jasper, H.; Kapahi, P. Dietary restriction improves intestinal cellular fitness to enhance gut barrier function and lifespan in D. melanogaster. PLoS Genet. 2018, 14, e1007777. [Google Scholar] [CrossRef]
- Hodge, B.A.; Meyerhof, G.T.; Katewa, S.D.; Lian, T.; Lau, C.; Bar, S.; Leung, N.Y.; Li, M.; Li-Kroeger, D.; Melov, S.; et al. Dietary restriction and the transcription factor clock delay eye aging to extend lifespan in Drosophila Melanogaster. Nat. Commun. 2022, 13, 3156. [Google Scholar] [CrossRef]
- Katewa, S.D.; Akagi, K.; Bose, N.; Rakshit, K.; Camarella, T.; Zheng, X.; Hall, D.; Davis, S.; Nelson, C.S.; Brem, R.B.; et al. Peripheral Circadian Clocks Mediate Dietary Restriction-Dependent Changes in Lifespan and Fat Metabolism in Drosophila. Cell Metab. 2016, 23, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Green, C.L.; Lamming, D.W.; Fontana, L. Molecular mechanisms of dietary restriction promoting health and longevity. Nat. Rev. Mol. Cell Biol. 2021, 23, 56–73. [Google Scholar] [CrossRef]
- Fontana, L.; Nehme, J.; Demaria, M. Caloric restriction and cellular senescence. Mech. Ageing Dev. 2018, 176, 19–23. [Google Scholar] [CrossRef]
- Belsky, D.W.; Caspi, A.; Corcoran, D.L.; Sugden, K.; Poulton, R.; Arseneault, L.; Baccarelli, A.; Chamarti, K.; Gao, X.; Hannon, E.; et al. DunedinPACE, a DNA methylation biomarker of the pace of aging. eLife 2022, 11, e73420. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, 3156. [Google Scholar] [CrossRef]
- Lu, A.T.; Quach, A.; Wilson, J.G.; Reiner, A.P.; Aviv, A.; Raj, K.; Hou, L.; Baccarelli, A.A.; Li, Y.; Stewart, J.D.; et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 2019, 11, 303–327. [Google Scholar] [CrossRef]
- Horvath, S.; Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 2018, 19, 371–384. [Google Scholar] [CrossRef]
- Petkovich, D.A.; Podolskiy, D.I.; Lobanov, A.V.; Lee, S.G.; Miller, R.A.; Gladyshev, V.N. Using DNA Methylation Profiling to Evaluate Biological Age and Longevity Interventions. Cell Metab. 2017, 25, 954–960.e956. [Google Scholar] [CrossRef] [PubMed]
- Poganik, J.R.; Zhang, B.; Baht, G.S.; Tyshkovskiy, A.; Deik, A.; Kerepesi, C.; Yim, S.H.; Lu, A.T.; Haghani, A.; Gong, T.; et al. Biological age is increased by stress and restored upon recovery. Cell Metab. 2023, 35, 807–820.e805. [Google Scholar] [CrossRef] [PubMed]
- Kemoun, P.; Ader, I.; Planat-Benard, V.; Dray, C.; Fazilleau, N.; Monsarrat, P.; Cousin, B.; Paupert, J.; Ousset, M.; Lorsignol, A.; et al. A gerophysiology perspective on healthy ageing. Ageing Res. Rev. 2022, 73, 101537. [Google Scholar] [CrossRef] [PubMed]
- Scheffer, M.; Bolhuis, J.E.; Borsboom, D.; Buchman, T.G.; Gijzel, S.M.W.; Goulson, D.; Kammenga, J.E.; Kemp, B.; van de Leemput, I.A.; Levin, S.; et al. Quantifying resilience of humans and other animals. Proc. Natl. Acad. Sci. USA 2018, 115, 11883–11890. [Google Scholar] [CrossRef]
- Cannon, W.B. Organization for Physiological Homeostasis. Physiol. Rev. 1929, 9, 399–431. [Google Scholar] [CrossRef]
- McEwen, B.S. Protective and damaging effects of stress mediators. N. Engl. J. Med. 1998, 338, 171–179. [Google Scholar] [CrossRef]
- Kallen, V.; Tahir, M.; Bedard, A.; Bongers, B.; van Riel, N.; van Meeteren, N. Aging and Allostasis: Using Bayesian Network Analytics to Explore and Evaluate Allostatic Markers in the Context of Aging. Diagnostics 2021, 11, 157. [Google Scholar] [CrossRef] [PubMed]
- Seeman, T.E.; McEwen, B.S.; Rowe, J.W.; Singer, B.H. Allostatic load as a marker of cumulative biological risk: MacArthur studies of successful aging. Proc. Natl. Acad. Sci. USA 2001, 98, 4770–4775. [Google Scholar] [CrossRef]
- Juster, R.P.; McEwen, B.S.; Lupien, S.J. Allostatic load biomarkers of chronic stress and impact on health and cognition. Neurosci. Biobehav. Rev. 2010, 35, 2–16. [Google Scholar] [CrossRef]
- McEwen, B.S.; Wingfield, J.C. The concept of allostasis in biology and biomedicine. Horm. Behav. 2003, 43, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Bobba-Alves, N.; Juster, R.P.; Picard, M. The energetic cost of allostasis and allostatic load. Psychoneuroendocrinology 2022, 146, 105951. [Google Scholar] [CrossRef] [PubMed]
- Bobba-Alves, N.; Sturm, G.; Lin, J.; Ware, S.A.; Karan, K.R.; Monzel, A.S.; Bris, C.; Procaccio, V.; Lenaers, G.; Higgins-Chen, A.; et al. Cellular allostatic load is linked to increased energy expenditure and accelerated biological aging. Psychoneuroendocrinology 2023, 155, 106322. [Google Scholar] [CrossRef]
- Mattson, M.P.; Arumugam, T.V. Hallmarks of Brain Aging: Adaptive and Pathological Modification by Metabolic States. Cell Metab. 2018, 27, 1176–1199. [Google Scholar] [CrossRef]
- Mattson, M.P.; Moehl, K.; Ghena, N.; Schmaedick, M.; Cheng, A. Intermittent metabolic switching, neuroplasticity and brain health. Nat. Rev. Neurosci. 2018, 19, 63–80. [Google Scholar] [CrossRef]
- Ferrucci, L.; Gonzalez-Freire, M.; Fabbri, E.; Simonsick, E.; Tanaka, T.; Moore, Z.; Salimi, S.; Sierra, F.; de Cabo, R. Measuring biological aging in humans: A quest. Aging Cell 2020, 19, e13080. [Google Scholar] [CrossRef]
- Chen, Z.; Raj, A.; Prateek, G.V.; Di Francesco, A.; Liu, J.; Keyes, B.E.; Kolumam, G.; Jojic, V.; Freund, A. Automated, high-dimensional evaluation of physiological aging and resilience in outbred mice. Elife 2022, 11, e72664. [Google Scholar] [CrossRef] [PubMed]
- Olde Rikkert, M.G.; Dakos, V.; Buchman, T.G.; Boer, R.; Glass, L.; Cramer, A.O.; Levin, S.; van Nes, E.; Sugihara, G.; Ferrari, M.D.; et al. Slowing Down of Recovery as Generic Risk Marker for Acute Severity Transitions in Chronic Diseases. Crit. Care Med. 2016, 44, 601–606. [Google Scholar] [CrossRef] [PubMed]
- Avchaciov, K.; Antoch, M.P.; Andrianova, E.L.; Tarkhov, A.E.; Menshikov, L.I.; Burmistrova, O.; Gudkov, A.V.; Fedichev, P.O. Unsupervised learning of aging principles from longitudinal data. Nat. Commun. 2022, 13, 6529. [Google Scholar] [CrossRef]
- Pyrkov, T.V.; Avchaciov, K.; Tarkhov, A.E.; Menshikov, L.I.; Gudkov, A.V.; Fedichev, P.O. Longitudinal analysis of blood markers reveals progressive loss of resilience and predicts human lifespan limit. Nat. Commun. 2021, 12, 2765. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.E.; Cropley, V.; Maier, A.B.; Lautenschlager, N.T.; Breakspear, M.; Zalesky, A. Heterogeneous aging across multiple organ systems and prediction of chronic disease and mortality. Nat. Med. 2023, 29, 1221–1231. [Google Scholar] [CrossRef]
- Tanaka, T.; Biancotto, A.; Moaddel, R.; Moore, A.Z.; Gonzalez-Freire, M.; Aon, M.A.; Candia, J.; Zhang, P.; Cheung, F.; Fantoni, G.; et al. Plasma proteomic signature of age in healthy humans. Aging Cell 2018, 17, e12799. [Google Scholar] [CrossRef]
- Fulop, T.; Larbi, A.; Dupuis, G.; Le Page, A.; Frost, E.H.; Cohen, A.A.; Witkowski, J.M.; Franceschi, C. Immunosenescence and Inflamm-Aging as Two Sides of the Same Coin: Friends or Foes? Front. Immunol. 2017, 8, 1960. [Google Scholar] [CrossRef]
- Franceschi, C.; Campisi, J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69 (Suppl. 1), S4–S9. [Google Scholar] [CrossRef]
- Fabbri, E.; An, Y.; Zoli, M.; Simonsick, E.M.; Guralnik, J.M.; Bandinelli, S.; Boyd, C.M.; Ferrucci, L. Aging and the burden of multimorbidity: Associations with inflammatory and anabolic hormonal biomarkers. J. Gerontol. A Biol. Sci. Med. Sci. 2015, 70, 63–70. [Google Scholar] [CrossRef] [PubMed]




| Models | Cell Types or Species | Datasets | References |
|---|---|---|---|
| Influenza A (H3N2) infection | Human | Microarray of the blood samples | [24,26] |
| COVID-19 infection | Human | Case reports in five different countries and regions | [25] |
| Hepatocellular carcinoma | Xenograft mouse model of HCCLM3 cells | Microarray of the liver samples | [27] |
| Breast cancer | Human breast adenocarcinoma MCF-7 cell line | RNA-seq of MCF-7 cells | [28] |
| Skin photodamage | The LSE model (3D skin model consisting of normal human keratinocyte and melanocyte) | RNA-seq of the LSE model | [29] |
| Lung cancer | KrasLSL-G12D/+; Lkb1flox/flox (KL) mice | RNA-seq of the KL lung samples | [30] |
| Hematopoietic stem cell differentiation | Mouse hematopoietic stem cells (mHSCs) | scRNA-seq of mHSCs | [31] |
| Embryonic stem cell differentiation | Human embryonic stem cells (hESCs) | scRNA-seq of hESCs | [32] |
| Immune cell differentiation | T cells from DO11.10 TCR mice | Raman imaging | [33] |
| Metabolic syndrome | Metabolic syndrome model mouse (TSOD mice) | Microarray of the adipose tissues | [34,35] |
| Type 2 diabetes | Diabetes model rat (GK rats) | Microarray of the adipose tissues | [36] |
| Senolytic Targets | Compound | References |
|---|---|---|
| SRC | Dasatinib | [76] |
| BCL-2 family | Quercetin, Navitoclax, A1331852, A1155463, Procyanidin C1 | [77,78,79] |
| HSP90 | Geldanamycin, Tanespimycin, 17-DMAG, Ansamycin, Resorcinol | [80,81] |
| PI3K | Fisetin, Luteolin, Enzastauin | [77,81] |
| p53-FOXO4 | FOXO4-DRI | [82] |
| Na+/K+ ATPase | Ouabain, Digoxin, Proscillaridin A, Bufalin | [83,84] |
| GLS1 | BPTES | [85] |
| Senomorphic Targets | Compound | References |
|---|---|---|
| mTOR, Nrf2, NF-κB | Rapamycin | [87,88] |
| NF-κB, Nrf2/GPx7, Insulin/IGF-1, mTOR etc. | Metformin | [89,90,91] |
| SIRT1 | Resveratrol, Sirtuin-activating compounds | [92,93,94,95] |
| NF-κB | SR12343 | [96] |
| p38MAPK | SB203580, UR13756, BIRB796 | [97,98] |
| JAK/STAT | Ruxolitinib | [99,100] |
| ATM | KU-55933, KU-60019 | [101,102] |
| HMG-CoA reductase | Atorvastatin, Pravastatin, Pitavastatin, Simvastatin | [103,104] |
| IRAK1/IκBα/NF-kB | Apigenin, Kaempferol | [105] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akagi, K.; Koizumi, K.; Kadowaki, M.; Kitajima, I.; Saito, S. New Possibilities for Evaluating the Development of Age-Related Pathologies Using the Dynamical Network Biomarkers Theory. Cells 2023, 12, 2297. https://doi.org/10.3390/cells12182297
Akagi K, Koizumi K, Kadowaki M, Kitajima I, Saito S. New Possibilities for Evaluating the Development of Age-Related Pathologies Using the Dynamical Network Biomarkers Theory. Cells. 2023; 12(18):2297. https://doi.org/10.3390/cells12182297
Chicago/Turabian StyleAkagi, Kazutaka, Keiichi Koizumi, Makoto Kadowaki, Isao Kitajima, and Shigeru Saito. 2023. "New Possibilities for Evaluating the Development of Age-Related Pathologies Using the Dynamical Network Biomarkers Theory" Cells 12, no. 18: 2297. https://doi.org/10.3390/cells12182297