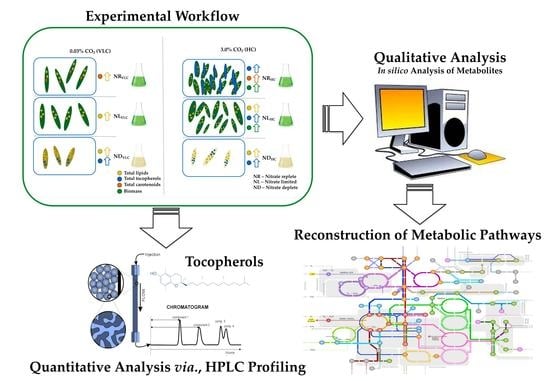
Multi-Fold Enhancement of Tocopherol Yields Employing High CO2 Supplementation and Nitrate Limitation in Native Isolate Monoraphidium sp.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Growth Conditions
2.2. Chlorophyll (Chla) Fluorescence Measurement
2.3. Biochemical Profiling
2.4. Confocal Microscopy with BODIPY Dye
2.5. Imaging of Superoxide Anion by Confocal Laser-Scanning Microscopy
2.6. Quantification of Tocopherols and Carotenoids by High-Performance Liquid Chromatography (HPLC)
2.7. Analysis of Total Antioxidant Activity
2.8. Qualitative (Untargeted) Metabolomics
2.9. Statistical Analysis
3. Results
3.1. Effect of Carbon Supplementation with Varying Nitrate Concentrations on Their Growth Profiles in the Native Isolate Monoraphidium sp. CABeR41
3.2. Changes in Biochemical Profiles under the Influence of Carbon Supplementation with Varying Nitrate Concentrations
3.3. Visualization of Lipid Droplets by BODIPY Staining
3.4. Imaging of Superoxide Anion Acitivity Employing Scanning Microscopy
3.5. Quantification of Tocopherols and Carotenoids in the Native Isolate Monoraphidium sp. Subjected to Carbon Supplementation with Varying Nitrate Concentrations
3.6. Analysis of Total Antioxidant Activity
3.7. Qualitative (Untargeted) Metabolomics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Cheng, J.; Zhu, Y.; Zhang, Z.; Yang, W. Modification and improvement of microalgae strains for strengthening CO2 fixation from coal-fired flue gas in power plants. Bioresour. Technol. 2019, 291, 121850. [Google Scholar] [CrossRef] [PubMed]
- Breuer, G.; Lamers, P.P.; Martens, D.E.; Draaisma, R.B.; Wijffels, R.H. The impact of nitrogen starvation on the dynamics of triacylglycerol accumulation in nine microalgae strains. Bioresour. Technol. 2012, 124, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Markou, G.; Georgakakis, D. Cultivation of filamentous cyanobacteria (blue-green algae) in agro-industrial wastes and wastewaters: A review. Appl. Energy 2011, 88, 3389–3401. [Google Scholar] [CrossRef]
- Rawat, I.; Ranjith Kumar, R.; Mutanda, T.; Bux, F. Dual role of microalgae: Phycoremediation of domestic wastewater and biomass production for sustainable biofuels production. Appl. Energy 2011, 88, 3411–3424. [Google Scholar] [CrossRef]
- Wu, M.; Gao, G.; Jian, Y.; Xu, J. High CO2 increases lipid and polyunsaturated fatty acid productivity of the marine diatom Skeletonema costatum in a two-stage model. J. Appl. Phycol. 2022, 34, 43–50. [Google Scholar] [CrossRef]
- Pospíšil, P. Molecular mechanisms of production and scavenging of reactive oxygen species by photosystem II. Biochim. Biophys. Acta-Bioenergy 2012, 1817, 218–231. [Google Scholar] [CrossRef] [Green Version]
- Fischer, B.B.; Hideg, É.; Krieger-Liszkay, A. Production, detection, and signaling of singlet oxygen in photosynthetic organisms. Antioxid. Redox Signal. 2013, 18, 2145–2162. [Google Scholar] [CrossRef]
- Kumari, P.; Cna’ani, A.; Didi-Cohen, S.; Tzin, V.; Khozin-Goldberg, I. Nitrogen deprivation-induced production of volatile organic compounds in the arachidonic-acid-accumulating microalga Lobosphaera incisa underpins their role as ros scavengers and chemical messengers. Front. Mar. Sci. 2020, 7, 410. [Google Scholar] [CrossRef]
- Sun, H.; Ren, Y.; Mao, X.; Li, X.; Zhang, H.; Lao, Y.; Chen, F. Harnessing C/N balance of Chromochloris zofingiensis to overcome the potential conflict in microalgal production. Commun. Biol. 2020, 3, 186. [Google Scholar] [CrossRef] [Green Version]
- Niyogi, K.K. Photoprotection revisited: Genetic and molecular approaches. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 333–359. [Google Scholar] [CrossRef]
- Rossel, J.B.; Wilson, I.W.; Pogson, B.J. Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol. 2002, 130, 1109–1120. [Google Scholar] [CrossRef] [Green Version]
- Müller-Moulé, P.; Havaux, M.; Niyogi, K.K. Zeaxanthin deficiency enhances the high light sensitivity of an ascorbate-deficient mutant of Arabidopsis. Plant Physiol. 2003, 133, 748–760. [Google Scholar] [CrossRef] [Green Version]
- Dörmann, P. Functional diversity of tocochromanols in plants. Planta 2007, 225, 269–276. [Google Scholar] [CrossRef]
- Li, Z.; Keasling, J.D.; Niyogi, K.K. Overlapping photoprotective function of vitamin e and carotenoids in Chlamydomonas. Plant Physiol. 2011, 158, 313–323. [Google Scholar] [CrossRef] [Green Version]
- Havaux, M.; Bonfils, J.P.; Lütz, C.; Niyogi, K.K. Photodamage of the photosynthetic apparatus and its dependence on the leaf developmental stage in the npq1 Arabidopsis mutant deficient in the xanthophyll cycle enzyme violaxanthin de-epoxidase. Plant Physiol. 2000, 124, 273–284. [Google Scholar] [CrossRef] [Green Version]
- Havaux, M.; Eymery, F.O.; Porfirova, S.; Rey, P.; Dörmann, P. Vitamin E protects against photoinhibition and photooxidative stress in Arabidopsis thaliana. Plant Cell 2005, 17, 3451–3469. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, A.; Vijayakumar, S.; Raman, K.; Srivastava, S. Rational metabolic engineering for enhanced alpha-tocopherol production in Helianthus annuus cell culture. Biochem. Eng. J. 2019, 151, 107256. [Google Scholar] [CrossRef]
- Lall, S.P.; Lewis-McCrea, L.M. Role of nutrients in skeletal metabolism and pathology in fish—An overview. Aquaculture 2007, 267, 3–19. [Google Scholar] [CrossRef]
- Brigelius-Flohé, R. Bioactivity of vitamin E. Nutr. Res. Rev. 2006, 19, 174–186. [Google Scholar] [CrossRef] [Green Version]
- Shintani, D.; DellaPenna, D. Elevating the vitamin E content of plants through metabolic engineering. Science 1998, 282, 2098–2100. [Google Scholar] [CrossRef]
- Velasco, L.; Fernández-Martínez, J.; Garcia-Ruiz, R. Genetic and environmental variation for tocopherol content and composition in sunflower commercial hybrids. J. Agric. Sci. 2002, 139, 425–429. [Google Scholar] [CrossRef]
- Hassapidou, M.N.; Balatsouras, G.D.; Manoukas, A.G. Effect of processing upon the tocopherol and tocotrienol composition of table olives. Food Chem. 1994, 50, 111–114. [Google Scholar] [CrossRef]
- Scherder, C.W.; Fehr, W.R.; Welke, G.A.; Wang, T. Tocopherol content and agronomic performance of soybean lines with reduced palmitate. Crop Sci. 2006, 46, 1286–1290. [Google Scholar] [CrossRef]
- Durmaz, Y. Vitamin E (α-tocopherol) production by the marine microalgae Nannochloropsis oculata (Eustigmatophyceae) in nitrogen limitation. Aquaculture 2007, 272, 717–722. [Google Scholar] [CrossRef]
- Ogbonna, J.C.; Tomiyamal, S.; Tanaka, H. Heterotrophic cultivation of Euglena gracilis Z for efficient production of α-tocopherol. J. Appl. Phycol. 1998, 10, 67–74. [Google Scholar] [CrossRef]
- Galasso, C.; Gentile, A.; Orefice, I.; Ianora, A.; Bruno, A.; Noonan, D.M.; Sansone, C.; Albini, A.; Brunet, C. Microalgal derivatives as potential nutraceutical and food supplements for human health: A focus on cancer prevention and interception. Nutrients 2019, 11, 1226. [Google Scholar] [CrossRef] [Green Version]
- Mudimu, O.; Rybalka, N.; Bauersachs, T.; Friedl, T.; Schulz, R. Influence of different CO2 concentrations on microalgae growth, α-tocopherol content and fatty acid composition. Geomicrobiol. J. 2015, 32, 291–303. [Google Scholar] [CrossRef]
- Fujita, T.; Aoyagi, H.; Ogbonna, J.C.; Tanaka, H. Effect of mixed organic substrate on alpha-tocopherol production by Euglena gracilis in photoheterotrophic culture. Appl. Microbiol. Biotechnol. 2008, 79, 371–378. [Google Scholar] [CrossRef]
- Afiukwa, C.; Ogbonna, J. Effects of mixed substrates on growth and vitamin production by Euglena gracilis. Afr. J. Biotechnol. 2007, 6, 2612–2615. [Google Scholar] [CrossRef]
- Mudimu, O.; Koopmann, I.K.; Rybalka, N.; Friedl, T.; Schulz, R.; Bilger, W. Screening of microalgae and cyanobacteria strains for α-tocopherol content at different growth phases and the influence of nitrate reduction on α-tocopherol production. J. Appl. Phycol. 2017, 29, 2867–2875. [Google Scholar] [CrossRef]
- Moheimani, N.R.; Borowitzka, M.A.; Isdepsky, A.; Sing, S.F. Standard methods for measuring growth of algae and their composition. In Algae for Biofuels and Energy; Borowitzka, M.A., Moheimani, N.R., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 265–284. [Google Scholar]
- Kareya, M.S.; Mariam, I.; Shaikh, K.M.; Nesamma, A.A.; Jutur, P.P. Photosynthetic carbon partitioning and metabolic regulation in response to very-low and high CO2 in Microchloropsis gaditana NIES 2587. Front. Plant Sci. 2020, 11, 981. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.; Harbinson, J.; Kramer, D. Determining the limitations and regulation of photosynthetic energy transduction in leaves. Plant Cell Environ. 2007, 30, 1107–1125. [Google Scholar] [CrossRef] [PubMed]
- Suggett, D.; Macintyre, H.; Kana, T.; Geider, R. Comparing electron transport with gas exchange: Parameterising exchange rates between alternative photosynthetic currencies for eukaryotic phytoplankton. Aquat. Microb. Ecol. 2009, 56, 147–162. [Google Scholar] [CrossRef] [Green Version]
- Mishra, S.K.; Suh, W.I.; Farooq, W.; Moon, M.; Shrivastav, A.; Park, M.S.; Yang, J.W. Rapid quantification of microalgal lipids in aqueous medium by a simple colorimetric method. Bioresour. Technol. 2014, 155, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Paliwal, C.; Pancha, I.; Ghosh, T.; Maurya, R.; Chokshi, K.; Vamsi Bharadwaj, S.V.; Ram, S.; Mishra, S. Selective carotenoid accumulation by varying nutrient media and salinity in Synechocystis sp. CCNM 2501. Bioresour. Technol. 2015, 197, 363–368. [Google Scholar] [CrossRef]
- Shaikh, K.M.; Nesamma, A.A.; Abdin, M.Z.; Jutur, P.P. Molecular profiling of an oleaginous trebouxiophycean alga Parachlorella kessleri subjected to nutrient deprivation for enhanced biofuel production. Biotechnol. Biofuels 2019, 12, 182. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; Gao, Z.; Li, F.; Fan, X.; Zhang, X.; Ye, N.; Mou, S.; Liang, C.; Li, D. Detection and quantitation of lipid in the microalga Tetraselmis subcordiformis (Wille) Butcher with BODIPY 505/515 staining. Bioresour. Technol. 2013, 127, 386–390. [Google Scholar] [CrossRef]
- Kumar, A.; Prasad, A.; Sedlářová, M.; Kale, R.; Frankel, L.K.; Sallans, L.; Bricker, T.M.; Pospíšil, P. Tocopherol controls D1 amino acid oxidation by oxygen radicals in photosystem II. Proc. Natl. Acad. Sci. USA 2021, 118, e2019246118. [Google Scholar] [CrossRef]
- Singh, R.; Paliwal, C.; Nesamma, A.A.; Narula, A.; Jutur, P.P. Nutrient deprivation mobilizes the production of unique tocopherols as a stress-promoting response in a new indigenous isolate Monoraphidium sp. Front. Mar. Sci. 2020, 7, 575817. [Google Scholar] [CrossRef]
- Paliwal, C.; Jutur, P.P. Dynamic allocation of carbon flux triggered by task-specific chemicals is an effective non-gene disruptive strategy for sustainable and cost-effective algal biorefineries. Chem. Eng. J. 2021, 418, 129413. [Google Scholar] [CrossRef]
- Burits, M.; Bucar, F. Antioxidant activity of Nigella sativa essential oil. Phytother. Res. 2000, 14, 323–328. [Google Scholar] [CrossRef]
- Paliwal, C.; Ghosh, T.; Bhayani, K.; Maurya, R.; Mishra, S. Antioxidant, anti-nephrolithe activities and in vitro digestibility studies of three different cyanobacterial pigment extracts. Mar. Drugs 2015, 13, 5384–5401. [Google Scholar] [CrossRef]
- Prieto, P.; Pineda, M.; Aguilar, M. Spectrophotometric quantitation of antioxidant capacity through the formation of a phosphomolybdenum complex: Specific application to the determination of vitamin E. Anal. Biochem. 1999, 269, 337–341. [Google Scholar] [CrossRef]
- Benzie, I.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [Green Version]
- Mariam, I.; Kareya, M.; Nesamma, A.; Jutur, P. Delineating metabolomic changes in native isolate Aurantiochytrium for production of docosahexaenoic acid in presence of varying carbon substrates. Algal Res. 2021, 55, 102285. [Google Scholar] [CrossRef]
- Minhas, A.K.; Hodgson, P.; Barrow, C.J.; Adholeya, A. A Review on the assessment of stress conditions for simultaneous production of microalgal lipids and carotenoids. Front. Microbiol. 2016, 7, 546. [Google Scholar] [CrossRef] [Green Version]
- Govindjee, S.A. On the relation between the Kautsky effect (chlorophyll a fluorescence induction) and photosystem II: Basics and applications of the OJIP fluorescence transient. Photochem. Photobiol. 2011, 104, 236–257. [Google Scholar] [CrossRef]
- Sarat Chandra, T.; Deepak, R.S.; Maneesh Kumar, M.; Mukherji, S.; Chauhan, V.S.; Sarada, R.; Mudliar, S.N. Evaluation of indigenous fresh water microalga Scenedesmus obtusus for feed and fuel applications: Effect of carbon dioxide, light and nutrient sources on growth and biochemical characteristics. Bioresour. Technol. 2016, 207, 430–439. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, H.P.; Han, B.; Yu, X. Coupling of abiotic stresses and phytohormones for the production of lipids and high-value by-products by microalgae: A review. Bioresour. Technol. 2019, 274, 549–556. [Google Scholar] [CrossRef]
- Gao, G.; Wu, M.; Fu, Q.; Li, X.; Xu, J. A two-stage model with nitrogen and silicon limitation enhances lipid productivity and biodiesel features of the marine bloom-forming diatom Skeletonema costatum. Bioresour. Technol. 2019, 289, 121717. [Google Scholar] [CrossRef]
- Jiang, X.; Han, Q.; Gao, X.; Gao, G. Conditions optimising on the yield of biomass, total lipid, and valuable fatty acids in two strains of Skeletonema menzelii. Food Chem. 2016, 194, 723–732. [Google Scholar] [CrossRef]
- Gordon, J.M.; Polle, J.E. Ultrahigh bioproductivity from algae. Appl. Microbiol. Biotechnol. 2007, 76, 969–975. [Google Scholar] [CrossRef]
- Schuhmann, H.; Lim, D.K.Y.; Schenk, P.M. Perspectives on metabolic engineering for increased lipid contents in microalgae. Biofuels 2012, 3, 71–86. [Google Scholar] [CrossRef]
- Kareya, M.S.; Mariam, I.; Rajacharya, G.H.; Nesamma, A.A.; Jutur, P.P. Valorization of carbon dioxide (CO2) to enhance production of biomass, biofuels, and biorenewables (B3) in Chlorella saccharophila UTEX247: A circular bioeconomy perspective. Biofuels Bioprod. Biorefin. 2021, 1–16. [Google Scholar] [CrossRef]
- Richardson, B.; Orcutt, D.M.; Schwertner, H.A.; Martinez, C.L.; Wickline, H.E. Effects of nitrogen limitation on the growth and composition of unicellular algae in continuous culture. Appl. Microbiol. 1969, 18, 245–250. [Google Scholar] [CrossRef]
- Lee, S.Y.; Hong, S.; Park, S.J.; Wegen, R.; Middelberg, A. Metabolic and kinetic analysis of poly(3-hydroxybutyrate) production by recombinant Escherichia coli. Biotechnol. Bioeng. 2001, 74, 70–80. [Google Scholar] [CrossRef]
- Sun, H.; Zhao, W.; Mao, X.; Li, Y.; Wu, T.; Chen, F. High-value biomass from microalgae production platforms: Strategies and progress based on carbon metabolism and energy conversion. Biotechnol. Biofuels 2018, 11, 227. [Google Scholar] [CrossRef] [Green Version]
- Murata, N.; Takahashi, S.; Nishiyama, Y.; Allakhverdiev, S.I. Photoinhibition of photosystem II under environmental stress. Biochim. Biophys. Acta-Bioenergy 2007, 1767, 414–421. [Google Scholar] [CrossRef] [Green Version]
- Mariam, I.; Kareya, M.S.; Rehmanji, M.; Nesamma, A.A.; Jutur, P.P. Channeling of carbon flux towards carotenogenesis in Botryococcus braunii: A media engineering perspective. Front. Microbiol. 2021, 12, 693106. [Google Scholar] [CrossRef]
- Zhao, L.-S.; Li, K.; Wang, Q.-M.; Song, X.-Y.; Su, H.-N.; Xie, B.-B.; Zhang, X.-Y.; Huang, F.; Chen, X.-L.; Zhou, B.-C.; et al. Nitrogen starvation impacts the photosynthetic performance of Porphyridium cruentum as revealed by chlorophyll a fluorescence. Sci. Rep. 2017, 7, 8542. [Google Scholar] [CrossRef]
- Triantaphylidès, C.; Krischke, M.; Hoeberichts, F.A.; Ksas, B.; Gresser, G.; Havaux, M.; Van Breusegem, F.; Mueller, M.J. Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiol. 2008, 148, 960–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auten, R.L.; Davis, J.M. Oxygen toxicity and reactive oxygen species: The devil is in the details. Pediatr. Res. 2009, 66, 121–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pospíšil, P. Production of reactive oxygen species by photosystem II. Biochim. Biophys. Acta 2009, 1787, 1151–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trebst, A.; Depka, B.; Holländer-Czytko, H. A specific role for tocopherol and of chemical singlet oxygen quenchers in the maintenance of photosystem II structure and function in Chlamydomonas reinhardtii. FEBS Lett. 2002, 516, 156–160. [Google Scholar] [CrossRef]
- Goiris, K.; Van Colen, W.; Wilches, I.; León-Tamariz, F.; De Cooman, L.; Muylaert, K. Impact of nutrient stress on antioxidant production in three species of microalgae. Algal Res. 2015, 7, 51–57. [Google Scholar] [CrossRef]
- Chen, G.; Wang, B.; Han, D.; Sommerfeld, M.; Lu, Y.; Chen, F.; Hu, Q. Molecular mechanisms of the coordination between astaxanthin and fatty acid biosynthesis in Haematococcus pluvialis (Chlorophyceae). Plant Mol. Biol. 2015, 81, 95–107. [Google Scholar] [CrossRef]
- Liu, J.; Mao, X.; Zhou, W.; Guarnieri, M.T. Simultaneous production of triacylglycerol and high-value carotenoids by the astaxanthin-producing oleaginous green microalga Chlorella zofingiensis. Bioresour. Technol. 2016, 214, 319–327. [Google Scholar] [CrossRef]
- Osorio, S.; Alba, R.; Damasceno, C.M.B.; Lopez-Casado, G.; Lohse, M.; Zanor, M.I.; Tohge, T.; Usadel, B.; Rose, J.K.C.; Fei, Z.; et al. Systems biology of tomato fruit development: Combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol. 2011, 157, 405–425. [Google Scholar] [CrossRef] [Green Version]
- Wase, N.; Black, P.N.; Stanley, B.A.; DiRusso, C.C. Integrated quantitative analysis of nitrogen stress response in Chlamydomonas reinhardtii using metabolite and protein profiling. J. Proteome Res. 2014, 13, 1373–1396. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics by gas chromatography-mass spectrometry: Combined targeted and untargeted profiling. Curr. Protoc. Mol. Biol. 2016, 114, 30.34.31–30.34.32. [Google Scholar] [CrossRef]
- Timm, S.; Florian, A.; Wittmiß, M.; Jahnke, K.; Hagemann, M.; Fernie, A.R.; Bauwe, H. Serine acts as a metabolic signal for the transcriptional control of photorespiration-related genes in Arabidopsis. Plant Physiol. 2013, 162, 379–389. [Google Scholar] [CrossRef] [Green Version]
- Dokulil, M.T. Phytoplankton productivity. In Reference Module in Earth Systems and Environmental Sciences; Elsevier Inc.: Amsterdam, The Netherlands, 2021. [Google Scholar] [CrossRef]
- Arora, N.; Dubey, D.; Sharma, M.; Patel, A.; Guleria, A.; Pruthi, P.A.; Kumar, D.; Pruthi, V.; Poluri, K.M. NMR-based metabolomic approach to elucidate the differential cellular responses during mitigation of arsenic (III, V) in a green microalga. ACS Omega 2018, 3, 11847–11856. [Google Scholar] [CrossRef]
- Martel, C.M. Nitrogen-deficient microalgae are rich in cell-surface mannose: Potential implications for prey biorecognition by phagotrophic protozoa. Braz. J. Microbiol. 2009, 40, 86–89. [Google Scholar] [CrossRef]
- Ubuka, T. Subchapter 132A—Glutamic acid. In Handbook of Hormones, 2nd ed.; Ando, H., Ukena, K., Nagata, S., Eds.; Academic Press: San Diego, CA, USA, 2021; pp. 1063–1065. [Google Scholar]
- Liu, J.; Sun, Z.; Mao, X.; Gerken, H.; Wang, X.; Yang, W. Multiomics analysis reveals a distinct mechanism of oleaginousness in the emerging model alga Chromochloris zofingiensis. Plant Mol. Biol. 2019, 98, 1060–1077. [Google Scholar] [CrossRef]
- Schmollinger, S.; Mühlhaus, T.; Boyle, N.R.; Blaby, I.K.; Casero, D.; Mettler, T.; Moseley, J.L.; Kropat, J.; Sommer, F.; Strenkert, D.; et al. Nitrogen-sparing mechanisms in Chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plant Cell 2014, 26, 1410–1435. [Google Scholar] [CrossRef] [Green Version]
- Guerra, L.T.; Levitan, O.; Frada, M.; Sun, J.; Falkowski, P.; Dismukes, G. Regulatory branch points affecting protein and lipid biosynthesis in the diatom Phaeodactylum tricornutum. Biomass Bioenergy 2013, 59, 306–315. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Cao, X.; Gao, P.; Liu, X.; Wang, X.; Zhang, J.; Zhou, J.; Xue, S.; Xu, G.; et al. Free amino acids and small molecular acids profiling of marine microalga Isochrysis zhangjiangensis under nitrogen deficiency. Algal Res. 2016, 13, 207–217. [Google Scholar] [CrossRef]
- Chen, H.; Zheng, Y.; Zhan, J.; He, C.; Wang, Q. Comparative metabolic profiling of the lipid-producing green microalga Chlorella reveals that nitrogen and carbon metabolic pathways contribute to lipid metabolism. Biotechnol. Biofuels 2017, 10, 153. [Google Scholar] [CrossRef] [Green Version]
- Sharma, N.; Chaudhary, C.; Khurana, P. Role of myo-inositol during skotomorphogenesis in Arabidopsis. Sci. Rep. 2020, 10, 17329. [Google Scholar] [CrossRef]
- Cho, K.; Kim, K.-N.; Lim, N.-L.; Kim, M.-S.; Ha, J.-C.; Shin, H.H.; Kim, M.-K.; Roh, S.W.; Kim, D.; Oda, T. Enhanced biomass and lipid production by supplement of myo-inositol with oceanic microalga Dunaliella salina. Biomass Bioenergy 2015, 72, 1–7. [Google Scholar] [CrossRef]
- Qiao, T.; Zhao, Y.; Han, B.; Li, T.; Zhao, P.; Xu, J.-W.; Huang, L.; Yu, X. Myo-inositol promotes lipid production and nutrients removal by microalga under molasses wastewater. Renew. Energy 2021, 172, 327–335. [Google Scholar] [CrossRef]
- Zhang, Y.; Adams, I.P.; Ratledge, C. Malic enzyme: The controlling activity for lipid production? Overexpression of malic enzyme in Mucor circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology 2007, 153, 2013–2025. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Min, Q.; Xu, J.; Zhang, K.; Chen, S.; Wang, H.; Li, D. Effect of malate on docosahexaenoic acid production from Schizochytrium sp. B4D1. Electron. J. Biotechnol. 2016, 19, 56–60. [Google Scholar] [CrossRef] [Green Version]
- Chia, D.W.; Yoder, T.J.; Reiter, W.-D.; Gibson, S.I. Fumaric acid: An overlooked form of fixed carbon in Arabidopsis and other plant species. Planta 2000, 211, 743–751. [Google Scholar] [CrossRef] [Green Version]
- Tschoep, H.; Gibon, Y.; Carillo, P.; Armengaud, P.; Szecowka, M.; Nunes-Nesi, A.; Fernie, A.R.; Koehl, K.; Stitt, M. Adjustment of growth and central metabolism to a mild but sustained nitrogen-limitation in Arabidopsis. Plant Cell. Environ. 2009, 32, 300–318. [Google Scholar] [CrossRef]
- Valenzuela, J.; Mazurie, A.; Carlson, R.P.; Gerlach, R.; Cooksey, K.E.; Peyton, B.M.; Fields, M.W. Potential role of multiple carbon fixation pathways during lipid accumulation in Phaeodactylum tricornutum. Biotechnol. Biofuels 2012, 5, 40. [Google Scholar] [CrossRef] [Green Version]
- Frayer, M.J. The antioxidant effects of thylakoid Vitamin E (α-tocopherol). Plant Cell Environ. 1992, 15, 381–392. [Google Scholar] [CrossRef]
- Munné-Bosch, S.; Alegre, L. The function of tocopherols and tocotrienols in plants. Crit. Rev. Plant Sci. 2002, 21, 31–57. [Google Scholar] [CrossRef]
- Vom Dorp, K.; Hölzl, G.; Plohmann, C.; Eisenhut, M.; Abraham, M.; Weber, A.P.M.; Hanson, A.D.; Dörmann, P. Remobilization of phytol from chlorophyll degradation is essential for tocopherol synthesis and growth of Aarabidopsis. Plant Cell 2015, 27, 2846–2859. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Parameters | NRVLC | NLVLC | NDVLC | NRHC | NLHC | NDHC |
|---|---|---|---|---|---|---|
| Biomass (g·L−1) | 0.71 ± 0.01 b | 0.65 ± 0.01 b | 0.38 ± 0.01 b | 3.54 ± 0.13 a | 3.30 ± 0.16 a | 0.62 ± 0.03 b |
| Biomass Productivity (mg·L−1 d−1) | 56.93 ± 0.76 b | 50.75 ± 0.89 b | 23.39 ± 1.14 b | 336.35 ± 14.92 a | 316.63 ± 15.58 a | 48.19 ± 2.96 b |
| Specific Growth Rate (µ) | 0.25 ± 0.02 | 0.19 ± 0.06 | 0.06 ± 0.03 | 0.44 ± 0.02 | 0.45 ± 0.01 | 0.12 ± 0.04 |
| Doubling Time (days) | 2.82 ± 0.18 | 3.58 ± 0.23 | 11.15 ± 0.86 | 1.60 ± 0.09 | 1.54 ± 0.04 | 6.50 ± 0.02 |
| Fv/Fm | 0.78 ± 0.01 | 0.55 ± 0.03 | 0.30 ± 0.09 | 0.80 ± 0.02 | 0.65 ± 0.06 | 0.00 ± 0.00 |
| Fq′/Fm′ | 0.45 ± 0.05 | 0.29 ± 0.07 | 0.15 ± 0.04 | 0.51 ± 0.05 | 0.36 ± 0.05 | 0.00 ± 0.00 |
| ETRII | 35.90 ± 3.54 | 23.35 ± 5.62 | 12.20 ± 3.46 | 41.10 ± 4.38 | 29.70 ± 5.16 | 0.00 ± 0.00 |
| Total Chlorophylls (mg·g−1 DCW) | 21.35 ± 1.92 b | 16.16 ± 1.36 b | 3.20 ± 0.54 cd | 42.53 ± 3.44 a | 15.02 ± 1.58 bc | 0.77 ± 0.18 d |
| Biochemical Constitutents | NRVLC | NLVLC | NDVLC | NRHC | NLHC | NDHC |
|---|---|---|---|---|---|---|
| Yields/Productivities (mg·g−1 DCW/mg·L−1 d−1) | ||||||
| Total Proteins | 508.76 ± 33.6/ 21.37 ± 1.7 c | 400.00 ± 21.0/ 16.73 ± 0.2 cd | 334.79 ± 67.6/ 0.53 ± 0.53 d | 660.43 ± 24.7/ 197.88 ± 6.5 a | 353.02 ± 34.1/ 79.47 ± 5.3 b | 201.70 ± 46.7/ 1.40 ± 0.5 d |
| Total Carbohydrates | 95.70 ± 3.4/ 5.24 ± 0.2 c | 70.37 ± 4.1/ 3.24 ± 0.3 c | 205.37 ± 10.4/ 6.10 ± 0.3 c | 106.64 ± 4.8/ 35.80 ± 1.3 b | 269.41 ± 26.5/ 87.12 ± 7.0 a | 177.15 ± 5.1/ 9.87 ± 0.5 c |
| Total Lipids | 114.00 ± 11.8/ 6.08 ± 0.3 c | 125.10 ± 21.0/ 7.50 ± 1.2 c | 193.15 ± 16.0/ 5.97 ± 0.3 c | 169.11 ± 17.5/ 49.68 ± 6.9 ab | 214.70 ± 29.8/ 76.12 ± 13.4 a | 344.70 ± 16.8/ 17.60 ± 3.3 bc |
| Tocopherols | NRVLC | NLVLC | NDVLC | NRHC | NLHC | NDHC |
|---|---|---|---|---|---|---|
| Yields/Productivities (μg.g−1 DCW/μg·L−1 d−1) | ||||||
| α-Tocopherol | 539.95 ± 69.2 bc/ 32.93 ± 4.9 | 425.01 ± 53.0 c/ 30.65 ± 2.9 | 468.20 ± 48.2 bc/ 19.45 ± 3.5 | 388.25 ± 3.9 c/ 137.07 ± 3.3 | 736.76 ± 22.5 ab/ 244.73 ± 7.4 | 947.72 ± 49.2 a/ 54.85 ± 6.5 |
| δ-Tocopherol | 528.10 ± 61.1 c/ 27.42 ± 2.4 | 687.50 ± 48.3 c/ 30.66 ± 3.1 | 925.50 ± 47.5 c/ 19.47 ± 0.7 | 478.20 ± 22.7 c/ 169.07 ± 0.4 | 1142.63 ± 159.6 b/ 489.65 ± 4.5 | 1795.31 ± 56.3 a/ 107.01 ± 5.4 |
| Total Tocopherols | 1068.04 ± 130.2/ 60.35 ± 7.3 de | 1111.51 ± 88.2/ 70.31 ± 8.2 d | 1393.16 ± 32.2/ 38.92 ± 4.2 e | 866.45 ± 26.7/ 306.14 ± 2.9 b | 1879.38 ± 137.5/ 734.38 ± 11.8 a | 2743.03 ± 7.21/ 161.85 ± 11.9 c |
| Carotenoids (in mg·g−1 DCW) | NRVLC | NLVLC | NDVLC | NRHC | NLHC | NDHC |
|---|---|---|---|---|---|---|
| Lycopene | 0.08 ± 0.02 a | 0.05 ± 0.01 a | 0.17 ± 0.03 a | 0.01 ± 0.00 a | 0.09 ± 0.00 a | 0.14 ± 0.00 a |
| α-Carotene | 0.22 ± 0.05 b | 0.19 ± 0.04 bc | 0.01 ± 0.00 c | 0.72 ± 0.02 a | 0.07 ± 0.02 bc | 0.00 ± 0.01 c |
| β-carotene | 0.30 ± 0.07 b | 0.20 ± 0.00 b | 0.11 ± 0.02 a | 0.49 ± 0.10 b | 0.22 ± 0.07 a | 0.03 ± 0.02 a |
| Zeaxanthin | 0.91 ± 0.05 b | 0.75 ± 0.07 bc | 0.45 ± 0.01 d | 1.43 ± 0.07 a | 0.62 ± 0.01 bc | 0.20 ± 0.01 c |
| Echinenone | 0.03 ± 0.01 b | 0.01 ± 0.00 b | 0.32 ± 0.03 a | 0.02 ± 0.00 b | 0.21 ± 0.04 a | 0.35 ± 0.07 a |
| Total carotenoids | 1.54 ± 0.08 b | 1.2 ± 0.05 bc | 1.06 ± 0.09 bc | 2.67 ± 0.15 a | 1.21 ± 0.21 bc | 0.72 ± 0.08 c |
| Assays (mg·g−1) | NRVLC | NLVLC | NDVLC | NRHC | NLHC | NDHC |
|---|---|---|---|---|---|---|
| TAC | 24.77 ± 4.02 a | 28.23 ± 4.08 a | 15.94 ± 1.88 b | 38.60 ± 9.23 a | 28.96 ± 7.32 a | 12.31 ± 2.17 b |
| FRAP | 11.08 ± 2.65 a | 10.13 ± 2.30 a | 10.70 ± 3.36 a | 13.30 ± 0.56 a | 13.36 ± 3.29 a | 5.07 ± 2.46 b |
| DPPH | 11.03 ± 4.77 b | 13.43 ± 6.57 b | 10.26 ± 9.75 c | 19.96 ± 5.00 a | 15.83 ± 4.30 b | 14.90 ± 4.28 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, R.; Nesamma, A.A.; Narula, A.; Jutur, P.P. Multi-Fold Enhancement of Tocopherol Yields Employing High CO2 Supplementation and Nitrate Limitation in Native Isolate Monoraphidium sp. Cells 2022, 11, 1315. https://doi.org/10.3390/cells11081315
Singh R, Nesamma AA, Narula A, Jutur PP. Multi-Fold Enhancement of Tocopherol Yields Employing High CO2 Supplementation and Nitrate Limitation in Native Isolate Monoraphidium sp. Cells. 2022; 11(8):1315. https://doi.org/10.3390/cells11081315
Chicago/Turabian StyleSingh, Rabinder, Asha Arumugam Nesamma, Alka Narula, and Pannaga Pavan Jutur. 2022. "Multi-Fold Enhancement of Tocopherol Yields Employing High CO2 Supplementation and Nitrate Limitation in Native Isolate Monoraphidium sp." Cells 11, no. 8: 1315. https://doi.org/10.3390/cells11081315