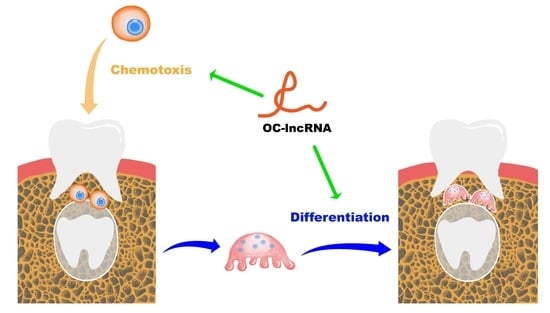
A Novel lncRNA Mediates the Delayed Tooth Eruption of Cleidocranial Dysplasia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Cell Culture and Osteoclast Induction
2.3. Total RNA Isolation and Real-Time PCR
2.4. RNA-seq and Bioinformatics Analysis
2.5. FISH Assays
2.6. Luciferase Assay
2.7. RNA Pull-Down Assay
2.8. Western Blot Analyses
2.9. TRAP Staining
2.10. Bone Resorption Assay
2.11. Immunohistochemistry
2.12. ELISA
2.13. Transient Transfection
2.14. Transwell Assay
2.15. RACE Assay
2.16. LncRNA Knock-Out by CRISPR/Cas9 Technique
2.17. Statistical Analysis
3. Results
3.1. Delayed Eruption of Permanent Teeth in CCD Primarily Results from Deficient Alveolar Bone Resorption
3.2. A Novel Osteoclast-Specific lncRNA Positively Regulates Osteoclast Differentiation and Bone Resorption
3.3. OC-lncRNA Positively Regulates Osteoclasts via a Competing Endogenous (ce)RNA Mechanism
3.4. CXCR3 Is a Positive Regulatory Gene in Osteoclast Differentiation and Bone Resorption
3.5. OC-lncRNA Reduction Diminishes Monocyte Chemotaxis through CXCR3 Downregulation
3.6. OC-lncRNA Facilitates Osteoclast Differentiation through CXCL10–CXCR3 Autocrine Signaling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Golan, I.; Baumert, U.; Hrala, B.P.; Mussig, D. Dentomaxillofacial variability of cleidocranial dysplasia: Clinicoradiological presentation and systematic review. Dentomaxillofac. Radiol. 2003, 32, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Impellizzeri, A.; Midulla, G.; Romeo, U.; La Monaca, C.; Barbato, E.; Galluccio, G. Delayed Eruption of Permanent Dentition and Maxillary Contraction in Patients with Cleidocranial Dysplasia: Review and Report of a Family. Int. J. Dent. 2018, 2018, 6591414. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zheng, S.; Wang, Y.; Zhao, Y.; Zhu, J.; Ge, L. Mutational analysis of RUNX2 gene in Chinese patients with cleidocranial dysplasia. Mutagenesis 2010, 25, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, Y.; Wang, X.; Sun, X.; Zhang, C.; Zheng, S. Analysis of novel RUNX2 mutations in Chinese patients with cleidocranial dysplasia. PLoS ONE 2017, 12, e0181653. [Google Scholar] [CrossRef]
- Wise, G.E.; Frazier-Bowers, S.; D’Souza, R.N. Cellular, molecular, and genetic determinants of tooth eruption. Crit. Rev. Oral Biol. Med. 2002, 13, 323–334. [Google Scholar] [CrossRef]
- Marks, S.C., Jr.; Cahill, D.R. Experimental study in the dog of the non-active role of the tooth in the eruptive process. Arch. Oral Biol. 1984, 29, 311–322. [Google Scholar] [CrossRef]
- Dorotheou, D.; Gkantidis, N.; Karamolegkou, M.; Kalyvas, D.; Kiliaridis, S.; Kitraki, E. Tooth eruption: Altered gene expression in the dental follicle of patients with cleidocranial dysplasia. Orthod. Craniofac. Res. 2013, 16, 20–27. [Google Scholar] [CrossRef]
- Chacon, G.E.; Ugalde, C.M.; Jabero, M.F. Genetic disorders and bone affecting the craniofacial skeleton. Oral Maxillofac. Surg. Clin. N. Am. 2007, 19, 467–474. [Google Scholar] [CrossRef]
- Xin, Y.; Liu, Y.; Liu, D.; Li, J.; Zhang, C.; Wang, Y.; Zheng, S. New Function of RUNX2 in Regulating Osteoclast Differentiation via the AKT/NFATc1/CTSK Axis. Calcif. Tissue Int. 2020, 106, 553–566. [Google Scholar] [CrossRef]
- Li, J.; Sarosi, I.; Yan, X.Q.; Morony, S.; Capparelli, C.; Tan, H.L.; McCabe, S.; Elliott, R.; Scully, S.; Van, G.; et al. RANK is the intrinsic hematopoietic cell surface receptor that controls osteoclastogenesis and regulation of bone mass and calcium metabolism. Proc. Natl. Acad. Sci. USA 2000, 97, 1566–1571. [Google Scholar] [CrossRef] [Green Version]
- Scimeca, J.C.; Franchi, A.; Trojani, C.; Parrinello, H.; Grosgeorge, J.; Robert, C.; Jaillon, O.; Poirier, C.; Gaudray, P.; Carle, G.F. The gene encoding the mouse homologue of the human osteoclast-specific 116-kDa V-ATPase subunit bears a deletion in osteosclerotic (oc/oc) mutants. Bone 2000, 26, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Yoda, S.; Suda, N.; Kitahara, Y.; Komori, T.; Ohyama, K. Delayed tooth eruption and suppressed osteoclast number in the eruption pathway of heterozygous Runx2/Cbfa1 knockout mice. Arch. Oral Biol. 2004, 49, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Proffit, W.R.; Frazier-Bowers, S.A. Mechanism and control of tooth eruption: Overview and clinical implications. Orthod. Craniofac. Res. 2009, 12, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Takarada, T.; Nakazato, R.; Tsuchikane, A.; Fujikawa, K.; Iezaki, T.; Yoneda, Y.; Hinoi, E. Genetic analysis of Runx2 function during intramembranous ossification. Development 2016, 143, 211–218. [Google Scholar] [CrossRef]
- Komori, T. Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 2010, 339, 189–195. [Google Scholar] [CrossRef]
- Ott, C.E.; Leschik, G.; Trotier, F.; Brueton, L.; Brunner, H.G.; Brussel, W.; Guillen-Navarro, E.; Haase, C.; Kohlhase, J.; Kotzot, D.; et al. Deletions of the RUNX2 gene are present in about 10% of individuals with cleidocranial dysplasia. Hum. Mutat. 2010, 31, E1587–E1593. [Google Scholar] [CrossRef]
- Wise, G.E.; Lumpkin, S.J.; Huang, H.; Zhang, Q. Osteoprotegerin and osteoclast differentiation factor in tooth eruption. J. Dent. Res. 2000, 79, 1937–1942. [Google Scholar] [CrossRef]
- Enomoto, H.; Shiojiri, S.; Hoshi, K.; Furuichi, T.; Fukuyama, R.; Yoshida, C.A.; Kanatani, N.; Nakamura, R.; Mizuno, A.; Zanma, A.; et al. Induction of osteoclast differentiation by Runx2 through receptor activator of nuclear factor-kappa B ligand (RANKL) and osteoprotegerin regulation and partial rescue of osteoclastogenesis in Runx2-/- mice by RANKL transgene. J. Biol. Chem. 2003, 278, 23971–23977. [Google Scholar] [CrossRef]
- Wise, G.E.; Yao, S. Regional differences of expression of bone morphogenetic protein-2 and RANKL in the rat dental follicle. Eur. J. Oral Sci. 2006, 114, 512–516. [Google Scholar] [CrossRef]
- Mundlos, S. Cleidocranial dysplasia: Clinical and molecular genetics. J. Med. Genet. 1999, 36, 177–182. [Google Scholar]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, M.; Li, X.L.; Hara, T.; Lal, A. A biochemical approach to identify direct microRNA targets. Methods Mol. Biol. 2015, 1206, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Takatsuna, H.; Asagiri, M.; Kubota, T.; Oka, K.; Osada, T.; Sugiyama, C.; Saito, H.; Aoki, K.; Ohya, K.; Takayanagi, H.; et al. Inhibition of RANKL-induced osteoclastogenesis by (-)-DHMEQ, a novel NF-kappaB inhibitor, through downregulation of NFATc1. J. Bone Miner. Res. 2005, 20, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.J.; Kim, B.; Kim, D.; Park Choo, H.Y.; Kim, H.H.; Ha, H.; Lee, Z.H. NF-kappaB signaling regulates cell-autonomous regulation of CXCL10 in breast cancer 4T1 cells. Exp. Mol. Med. 2017, 49, e295. [Google Scholar] [CrossRef]
- Wise, G.E.; King, G.J. Mechanisms of tooth eruption and orthodontic tooth movement. J. Dent. Res. 2008, 87, 414–434. [Google Scholar] [CrossRef]
- Cahill, D.R. Eruption pathway formation in the presence of experimental tooth impaction in puppies. Anat. Rec. 1969, 164, 67–77. [Google Scholar] [CrossRef]
- Sharma, S.; Mahajan, A.; Mittal, A.; Gohil, R.; Sachdeva, S.; Khan, S.; Dhillon, M. Epigenetic and transcriptional regulation of osteoclastogenesis in the pathogenesis of skeletal diseases: A systematic review. Bone 2020, 138, 115507. [Google Scholar] [CrossRef]
- Nagano, T.; Fraser, P. No-nonsense functions for long noncoding RNAs. Cell 2011, 145, 178–181. [Google Scholar] [CrossRef]
- Amaral, P.P.; Mattick, J.S. Noncoding RNA in development. Mamm. Genome 2008, 19, 454–492. [Google Scholar] [CrossRef]
- Liu, W.; Li, Z.; Cai, Z.; Xie, Z.; Li, J.; Li, M.; Cen, S.; Tang, S.; Zheng, G.; Ye, G.; et al. LncRNA-mRNA expression profiles and functional networks in osteoclast differentiation. J. Cell Mol. Med. 2020, 24, 9786–9797. [Google Scholar] [CrossRef]
- Dou, C.; Cao, Z.; Yang, B.; Ding, N.; Hou, T.; Luo, F.; Kang, F.; Li, J.; Yang, X.; Jiang, H.; et al. Changing expression profiles of lncRNAs, mRNAs, circRNAs and miRNAs during osteoclastogenesis. Sci. Rep. 2016, 6, 21499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Fu, S.; Sun, D.; Xing, J.; Hou, T.; Wu, X. EPC-derived exosomes promote osteoclastogenesis through LncRNA-MALAT1. J. Cell Mol. Med. 2019, 23, 3843–3854. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.; Hu, H.L.; Liu, K.Y.; Ram, Y.I.; Gao, J.L.; Cao, Y.M. Long noncoding RNA MIRG induces osteoclastogenesis and bone resorption in osteoporosis through negative regulation of miR-1897. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10195–10203. [Google Scholar] [CrossRef] [PubMed]
- Lacotte, S.; Brun, S.; Muller, S.; Dumortier, H. CXCR3, inflammation, and autoimmune diseases. Ann. N. Y. Acad. Sci. 2009, 1173, 310–317. [Google Scholar] [CrossRef]
- Cannon, A.; Thompson, C.M.; Maurer, H.C.; Atri, P.; Bhatia, R.; West, S.; Ghersi, D.; Olive, K.P.; Kumar, S.; Batra, S.K. CXCR3 and Cognate Ligands are Associated with Immune Cell Alteration and Aggressiveness of Pancreatic Ductal Adenocarcinoma. Clin. Cancer Res. 2020, 26, 6051–6063. [Google Scholar] [CrossRef]
- Guo, Y.; Kasahara, S.; Jhingran, A.; Tosini, N.L.; Zhai, B.; Aufiero, M.A.; Mills, K.A.M.; Gjonbalaj, M.; Espinosa, V.; Rivera, A.; et al. During Aspergillus Infection, Monocyte-Derived DCs, Neutrophils, and Plasmacytoid DCs Enhance Innate Immune Defense through CXCR3-Dependent Crosstalk. Cell Host Microbe 2020, 28, 104–116 e104. [Google Scholar] [CrossRef]
- Maurice, N.J.; McElrath, M.J.; Andersen-Nissen, E.; Frahm, N.; Prlic, M. CXCR3 enables recruitment and site-specific bystander activation of memory CD8(+) T cells. Nat. Commun. 2019, 10, 4987. [Google Scholar] [CrossRef]
- Kwak, H.B.; Ha, H.; Kim, H.N.; Lee, J.H.; Kim, H.S.; Lee, S.; Kim, H.M.; Kim, J.Y.; Kim, H.H.; Song, Y.W.; et al. Reciprocal cross-talk between RANKL and interferon-gamma-inducible protein 10 is responsible for bone-erosive experimental arthritis. Arthritis Rheum. 2008, 58, 1332–1342. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, H.N.; Kim, K.O.; Jin, W.J.; Lee, S.; Kim, H.H.; Ha, H.; Lee, Z.H. CXCL10 promotes osteolytic bone metastasis by enhancing cancer outgrowth and osteoclastogenesis. Cancer Res. 2012, 72, 3175–3186. [Google Scholar] [CrossRef]
- Jimi, E.; Aoki, K.; Saito, H.; D’Acquisto, F.; May, M.J.; Nakamura, I.; Sudo, T.; Kojima, T.; Okamoto, F.; Fukushima, H.; et al. Selective inhibition of NF-kappa B blocks osteoclastogenesis and prevents inflammatory bone destruction in vivo. Nat. Med. 2004, 10, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Abu-Amer, Y. IL-4 abrogates osteoclastogenesis through STAT6-dependent inhibition of NF-kappaB. J. Clin. Investig. 2001, 107, 1375–1385. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Lee, J.H.; Jin, W.J.; Kim, H.H.; Ha, H.; Lee, Z.H. JN-2, a C-X-C motif chemokine receptor 3 antagonist, ameliorates arthritis progression in an animal model. Eur. J. Pharmacol. 2018, 823, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, B.; Ma, Y.; Yi, J.; Zhang, C.; Zhang, Y.; Xu, Z.; Wang, J.; Yang, K.; Yang, A.; et al. Hantaan virus infection induces CXCL10 expression through TLR3, RIG-I, and MDA-5 pathways correlated with the disease severity. Mediat. Inflamm. 2014, 2014, 697837. [Google Scholar] [CrossRef]
- Shin, S.Y.; Nam, J.S.; Lim, Y.; Lee, Y.H. TNFalpha-exposed bone marrow-derived mesenchymal stem cells promote locomotion of MDA-MB-231 breast cancer cells through transcriptional activation of CXCR3 ligand chemokines. J. Biol. Chem. 2010, 285, 30731–30740. [Google Scholar] [CrossRef]
- Lv, Z.T.; Liang, S.; Chen, K.; Zhang, J.M.; Cheng, P.; Guo, J.C.; Yang, Q.; Zhou, C.H.; Liao, H.; Chen, A.M. FNDC4 Inhibits RANKL-Induced Osteoclast Formation by Suppressing NF-kappaB Activation and CXCL10 Expression. BioMed Res. Int. 2018, 2018, 3936257. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, B.; Jin, W.J.; Kim, H.H.; Ha, H.; Lee, Z.H. Pathogenic roles of CXCL10 signaling through CXCR3 and TLR4 in macrophages and T cells: Relevance for arthritis. Arthritis Res. Ther. 2017, 19, 163. [Google Scholar] [CrossRef]







| Patient | Sex | Age (Years) | Cranial Sign | Clavicular Sign | Delayed Eruption of Permanent Teeth (Number) | Supernumerary Teeth | Mutation |
|---|---|---|---|---|---|---|---|
| #1 | Male | 25 | Yes | Yes | Yes (16) | Yes | c.644delG |
| #2 | Female | 26 | Yes | Yes | Yes (19) | No | c.674G > T |
| #3 | Female | 26 | Yes | Yes | Yes (7) | Yes | c.559C > T |
| #4 | Male | 29 | Yes | Yes | Yes (18) | Yes | c.569G > A |
| #5 | Male | 20 | Yes | Yes | Yes (14) | Yes | c. 514delT a |
| #6 | Female | 23 | Yes | Yes | Yes (14) | No | — |
| #7 | Male | 16 | Yes | Yes | Yes (25) | Yes | c.673C > T |
| #8 | Female | 18 | Yes | Yes | Yes (8) | Yes | c.199C > T |
| #9 | Female | 46 | Yes | Yes | Yes (7) | Yes | c.557G > C |
| #10 | Male | 20 | Yes | Yes | Yes (6) | Yes | c.338T > G |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xin, Y.; Liu, Y.; Li, J.; Liu, D.; Zhang, C.; Wang, Y.; Zheng, S. A Novel lncRNA Mediates the Delayed Tooth Eruption of Cleidocranial Dysplasia. Cells 2022, 11, 2729. https://doi.org/10.3390/cells11172729
Xin Y, Liu Y, Li J, Liu D, Zhang C, Wang Y, Zheng S. A Novel lncRNA Mediates the Delayed Tooth Eruption of Cleidocranial Dysplasia. Cells. 2022; 11(17):2729. https://doi.org/10.3390/cells11172729
Chicago/Turabian StyleXin, Yuejiao, Yang Liu, Jie Li, Dandan Liu, Chenying Zhang, Yixiang Wang, and Shuguo Zheng. 2022. "A Novel lncRNA Mediates the Delayed Tooth Eruption of Cleidocranial Dysplasia" Cells 11, no. 17: 2729. https://doi.org/10.3390/cells11172729