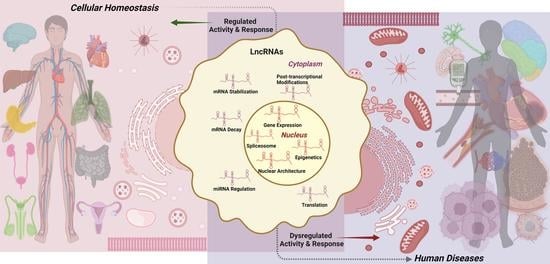
Signaling by LncRNAs: Structure, Cellular Homeostasis, and Disease Pathology
Abstract
:1. Introduction
2. Structural Features of lncRNAs
2.1. Primary Structure
2.2. Secondary Structure
2.3. Tertiary Structure
3. LncRNAs: Functional Mechanisms
3.1. Nuclear Functions
3.1.1. Nuclear Architecture
3.1.2. Dosage Compensation and Genomic Imprinting
3.1.3. Epigenetic Regulation
3.1.4. Regulation of Transcription
3.2. Cytosolic Functions
3.2.1. Regulation of mRNA Stability and Translation
3.2.2. Sequestration of miRNAs
3.2.3. Regulation of lncRNA-Specific Interactomes
4. LncRNAs in Human Diseases
4.1. LncRNAs in Cardiovascular Diseases
4.1.1. LncRNAs in Arterial Hypertension
4.1.2. LncRNAs in Coronary Heart Diseases
4.1.3. LncRNAs in Acute Myocardial Infarction
4.1.4. LncRNAs in Heart Failure
4.2. LncRNAs in Neurological Disorders
4.2.1. LncRNAs in Alzheimer’s Disease
4.2.2. LncRNAs in Parkinson’s Disease
4.2.3. LncRNAs in Amyotrophic Lateral Sclerosis
4.2.4. LncRNAs in Huntington’s Disease
4.2.5. LncRNAs in Autism Spectrum Disorder
4.2.6. LncRNAs in Schizophrenia
4.3. LncRNAs in Cancer
4.3.1. LncRNAs and Cell Proliferation
4.3.2. LncRNAs and Genomic Instability
4.3.3. LncRNAs and Metabolic Reprogramming
4.3.4. LncRNAs and Immune Evasion
4.3.5. LncRNAs and Epithelial–Mesenchymal Transition
4.3.6. LncRNAs and Tumor Angiogenesis
4.3.7. LncRNAs and Cancer Stemness
4.3.8. LncRNAs and Therapy Resistance
4.4. LncRNAs in Diabetes Mellitus
4.4.1. LncRNAs in Insulin Resistance and Synthesis
4.4.2. LncRNAs in Glucose Metabolism
4.4.3. LncRNAs in Diabetic Nephropathy
4.4.4. LncRNAs in Diabetic Retinopathy
4.4.5. LncRNAs in Diabetic Neuropathic Pain
4.4.6. LncRNAs in Diabetic Cardiac Myopathy
4.4.7. LncRNAs in Diabetes-Associated Inflammation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crick, F.H. On protein synthesis. Symp. Soc. Exp. Biol. 1958, 12, 138–163. [Google Scholar] [PubMed]
- Crick, F. Central dogma of molecular biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Baltimore, D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature 1970, 226, 1209–1211. [Google Scholar] [CrossRef] [PubMed]
- Temin, H.M.; Mizutani, S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 1970, 226, 1211–1213. [Google Scholar] [CrossRef] [PubMed]
- Saghatelian, A.; Couso, J.P. Discovery and characterization of smORF-encoded bioactive polypeptides. Nat. Chem. Biol. 2015, 11, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Frankish, A.; Diekhans, M.; Ferreira, A.-M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019, 47, D766–D773. [Google Scholar] [CrossRef]
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018, 46, D308–D314. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef]
- Balas, M.M.; Johnson, A.M. Exploring the mechanisms behind long noncoding RNAs and cancer. Noncoding RNA Res. 2018, 3, 108–117. [Google Scholar] [CrossRef]
- Carninci, P. Molecular biology: The long and short of RNAs. Nature 2009, 457, 974–975. [Google Scholar] [CrossRef] [PubMed]
- Kazimierczyk, M.; Kasprowicz, M.K.; Kasprzyk, M.E.; Wrzesinski, J. Human Long Noncoding RNA Interactome: Detection, Characterization and Function. Int. J. Mol. Sci. 2020, 21, 1027. [Google Scholar] [CrossRef] [PubMed]
- Losko, M.; Kotlinowski, J.; Jura, J. Long noncoding RNAs in metabolic syndrome related disorders. Mediat. Inflamm. 2016, 2016, 5365209. [Google Scholar] [CrossRef] [PubMed]
- Romero-Barrios, N.; Legascue, M.F.; Benhamed, M.; Ariel, F.; Crespi, M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018, 46, 2169–2184. [Google Scholar] [CrossRef]
- Chillón, I.; Marcia, M. The molecular structure of long non-coding RNAs: Emerging patterns and functional implications. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 662–690. [Google Scholar] [CrossRef]
- Brown, J.A.; Valenstein, M.L.; Yario, T.A.; Tycowski, K.T.; Steitz, J.A. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENβ noncoding RNAs. Proc. Natl. Acad. Sci. USA 2012, 109, 19202–19207. [Google Scholar] [CrossRef]
- Wilusz, J.E.; JnBaptiste, C.K.; Lu, L.Y.; Kuhn, C.-D.; Joshua-Tor, L.; Sharp, P.A. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly (A) tails. Genes Dev. 2012, 26, 2392–2407. [Google Scholar] [CrossRef]
- Chillón, I.; Pyle, A.M. Inverted repeat Alu elements in the human lincRNA-p21 adopt a conserved secondary structure that regulates RNA function. Nucleic Acids Res. 2016, 44, 9462–9471. [Google Scholar]
- Tassinari, M.; Richter, S.N.; Gandellini, P. Biological relevance and therapeutic potential of G-quadruplex structures in the human noncoding transcriptome. Nucleic Acids Res 2021, 49, 3617–3633. [Google Scholar] [CrossRef]
- Wu, L.; Murat, P.; Matak-Vinkovic, D.; Murrell, A.; Balasubramanian, S. Binding Interactions between Long Noncoding RNA HOTAIR and PRC2 Proteins. Biochemistry 2013, 52, 9519–9527. [Google Scholar] [CrossRef]
- Hirashima, K.; Seimiya, H. Telomeric repeat-containing RNA/G-quadruplex-forming sequences cause genome-wide alteration of gene expression in human cancer cells in vivo. Nucleic Acids Res. 2015, 43, 2022–2032. [Google Scholar] [CrossRef]
- Arora, R.; Lee, Y.; Wischnewski, H.; Brun, C.; Schwarz, T.; Azzalin, C. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014, 5, 5220. [Google Scholar] [CrossRef]
- Ergun, S.; Oztuzcu, S. Oncocers: ceRNA-mediated cross-talk by sponging miRNAs in oncogenic pathways. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2015, 36, 3129–3136. [Google Scholar] [CrossRef]
- Lubelsky, Y.; Ulitsky, I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature 2018, 555, 107–111. [Google Scholar] [CrossRef]
- Yin, Y.; Lu, J.Y.; Zhang, X.; Shao, W.; Xu, Y.; Li, P.; Hong, Y.; Cui, L.; Shan, G.; Tian, B. U1 snRNP regulates chromatin retention of noncoding RNAs. Nature 2020, 580, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Cruz, J.A.; Westhof, E. The dynamic landscapes of RNA architecture. Cell 2009, 136, 604–609. [Google Scholar] [CrossRef]
- Novikova, I.V.; Hennelly, S.P.; Tung, C.S.; Sanbonmatsu, K.Y. Rise of the RNA machines: Exploring the structure of long non-coding RNAs. J. Mol. Biol. 2013, 425, 3731–3746. [Google Scholar] [CrossRef]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef]
- Somarowthu, S.; Legiewicz, M.; Chillón, I.; Marcia, M.; Liu, F.; Pyle, A.M. HOTAIR forms an intricate and modular secondary structure. Mol. Cell 2015, 58, 353–361. [Google Scholar] [CrossRef]
- Uroda, T.; Anastasakou, E.; Rossi, A.; Teulon, J.-M.; Pellequer, J.-L.; Annibale, P.; Pessey, O.; Inga, A.; Chillón, I.; Marcia, M. Conserved pseudoknots in lncRNA MEG3 are essential for stimulation of the p53 pathway. Mol. Cell 2019, 75, 982–995.e9. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, E.J.; Hennelly, S.P.; Novikova, I.V.; Irwin, J.A.; Dean, C.; Sanbonmatsu, K.Y. COOLAIR antisense RNAs form evolutionarily conserved elaborate secondary structures. Cell Rep. 2016, 16, 3087–3096. [Google Scholar] [CrossRef] [PubMed]
- Tycowski, K.T.; Shu, M.D.; Borah, S.; Shi, M.; Steitz, J.A. Conservation of a triple-helix-forming RNA stability element in noncoding and genomic RNAs of diverse viruses. Cell Rep. 2012, 2, 26–32. [Google Scholar] [CrossRef]
- Mitton-Fry, R.M.; DeGregorio, S.J.; Wang, J.; Steitz, T.A.; Steitz, J.A. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science 2010, 330, 1244–1247. [Google Scholar] [CrossRef]
- Wilusz, J.E.; Spector, D.L. An unexpected ending: Noncanonical 3′ end processing mechanisms. RNA 2010, 16, 259–266. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010, 3, ra8. [Google Scholar] [CrossRef]
- Hudson, W.H.; Pickard, M.R.; De Vera, I.M.S.; Kuiper, E.G.; Mourtada-Maarabouni, M.; Conn, G.L.; Kojetin, D.J.; Williams, G.T.; Ortlund, E.A. Conserved sequence-specific lincRNA–steroid receptor interactions drive transcriptional repression and direct cell fate. Nat. Commun. 2014, 5, 5395. [Google Scholar] [CrossRef]
- Graf, J.; Kretz, M. From structure to function: Route to understanding lncRNA mechanism. Bioessays 2020, 42, e2000027. [Google Scholar] [CrossRef]
- Uroda, T.; Chillón, I.; Annibale, P.; Teulon, J.-M.; Pessey, O.; Karuppasamy, M.; Pellequer, J.-L.; Marcia, M. Visualizing the functional 3D shape and topography of long noncoding RNAs by single-particle atomic force microscopy and in-solution hydrodynamic techniques. Nat. Protoc. 2020, 15, 2107–2139. [Google Scholar] [CrossRef] [PubMed]
- Guh, C.-Y.; Hsieh, Y.-H.; Chu, H.-P. Functions and properties of nuclear lncRNAs—From systematically mapping the interactomes of lncRNAs. J. Biomed. Sci. 2020, 27, 44. [Google Scholar] [CrossRef] [PubMed]
- Spector, D.L.; Lamond, A.I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011, 3, a000646. [Google Scholar] [CrossRef]
- Miyagawa, R.; Tano, K.; Mizuno, R.; Nakamura, Y.; Ijiri, K.; Rakwal, R.; Shibato, J.; Masuo, Y.; Mayeda, A.; Hirose, T.; et al. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA 2012, 18, 738–751. [Google Scholar] [CrossRef]
- Fox, A.H.; Lamond, A.I. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2010, 2, a000687. [Google Scholar] [CrossRef]
- Sunwoo, H.; Dinger, M.E.; Wilusz, J.E.; Amaral, P.P.; Mattick, J.S.; Spector, D.L. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009, 19, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Hacisuleyman, E.; Goff, L.A.; Trapnell, C.; Williams, A.; Henao-Mejia, J.; Sun, L.; McClanahan, P.; Hendrickson, D.G.; Sauvageau, M.; Kelley, D.R.; et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014, 21, 198–206. [Google Scholar] [CrossRef]
- Trimarchi, T.; Bilal, E.; Ntziachristos, P.; Fabbri, G.; Dalla-Favera, R.; Tsirigos, A.; Aifantis, I. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell 2014, 158, 593–606. [Google Scholar] [CrossRef]
- Medyouf, H.; Gusscott, S.; Wang, H.; Tseng, J.C.; Wai, C.; Nemirovsky, O.; Trumpp, A.; Pflumio, F.; Carboni, J.; Gottardis, M.; et al. High-level IGF1R expression is required for leukemia-initiating cell activity in T-ALL and is supported by Notch signaling. J. Exp. Med. 2011, 208, 1809–1822. [Google Scholar] [CrossRef]
- Yan, F.; Wang, X.; Zeng, Y. 3D genomic regulation of lncRNA and Xist in X chromosome. In Seminars in Cell & Developmental Biology; Academic Press: Cambridge, MA, USA, 2019; pp. 174–180. [Google Scholar]
- McHugh, C.A.; Chen, C.K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef]
- Cerase, A.; Pintacuda, G.; Tattermusch, A.; Avner, P. Xist localization and function: New insights from multiple levels. Genome Biol. 2015, 16, 166. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Ishihara, K.; Kato, R. Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation. J. Biochem. 2000, 127, 711–715. [Google Scholar] [CrossRef] [PubMed]
- Monnier, P.; Martinet, C.; Pontis, J.; Stancheva, I.; Ait-Si-Ali, S.; Dandolo, L. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc. Natl. Acad. Sci. USA 2013, 110, 20693. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Yao, L.; Zhang, Q.; Wang, F.; Mei, H.; Guo, X.; Huang, W. Long noncoding RNA HOTAIR promotes metastasis of renal cell carcinoma by up-regulating histone H3K27 demethylase JMJD3. Oncotarget 2017, 8, 19795–19802. [Google Scholar] [CrossRef]
- Portoso, M.; Ragazzini, R.; Brenčič, Ž.; Moiani, A.; Michaud, A.; Vassilev, I.; Wassef, M.; Servant, N.; Sargueil, B.; Margueron, R. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J. 2017, 36, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Jarroux, J.; Foretek, D.; Bertrand, C.; Gabriel, M.; Szachnowski, U.; Saci, Z.; Guo, S.; Londoño-Vallejo, A.; Pinskaya, M.; Morillon, A. HOTAIR lncRNA promotes epithelial–mesenchymal transition by redistributing LSD1 at regulatory chromatin regions. EMBO Rep. 2021, 22, e50193. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Merry, C.R.; Forrest, M.E.; Sabers, J.N.; Beard, L.; Gao, X.H.; Hatzoglou, M.; Jackson, M.W.; Wang, Z.; Markowitz, S.D.; Khalil, A.M. DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer. Hum. Mol. Genet. 2015, 24, 6240–6253. [Google Scholar] [CrossRef]
- Somasundaram, S.; Forrest, M.E.; Moinova, H.; Cohen, A.; Varadan, V.; LaFramboise, T.; Markowitz, S.; Khalil, A.M. The DNMT1-associated lincRNA DACOR1 reprograms genome-wide DNA methylation in colon cancer. Clin. Epigenet. 2018, 10, 127. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Latos, P.A.; Pauler, F.M.; Koerner, M.V.; Şenergin, H.B.; Hudson, Q.J.; Stocsits, R.R.; Allhoff, W.; Stricker, S.H.; Klement, R.M.; Warczok, K.E.; et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 2012, 338, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Groff, A.F.; Sanchez-Gomez, D.B.; Soruco, M.M.L.; Gerhardinger, C.; Barutcu, A.R.; Li, E.; Elcavage, L.; Plana, O.; Sanchez, L.V.; Lee, J.C.; et al. In Vivo Characterization of Linc-p21 Reveals Functional cis-Regulatory DNA Elements. Cell Rep. 2016, 16, 2178–2186. [Google Scholar] [CrossRef] [PubMed]
- Vance, K.W.; Sansom, S.N.; Lee, S.; Chalei, V.; Kong, L.; Cooper, S.E.; Oliver, P.L.; Ponting, C.P. The long non-coding RNA Paupar regulates the expression of both local and distal genes. EMBO J. 2014, 33, 296–311. [Google Scholar] [CrossRef]
- Vance, K.W.; Ponting, C.P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014, 30, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Schoenberg, D.R.; Maquat, L.E. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 2012, 13, 246–259. [Google Scholar] [CrossRef]
- Gong, C.; Maquat, L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 2011, 470, 284–288. [Google Scholar] [CrossRef]
- Kretz, M.; Siprashvili, Z.; Chu, C.; Webster, D.E.; Zehnder, A.; Qu, K.; Lee, C.S.; Flockhart, R.J.; Groff, A.F.; Chow, J.; et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013, 493, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.p.; Liu, X.x.; Xia, R.; Yin, L.; Kong, R.; Chen, W.m.; Huang, M.d.; Shu, Y.q. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene 2015, 34, 5648–5661. [Google Scholar] [CrossRef]
- Li, Y.; Guo, D.; Ren, M.; Zhao, Y.; Wang, X.; Chen, Y.; Liu, Y.; Lu, G.; He, S. Long non-coding RNA SNAI3-AS1 promotes the proliferation and metastasis of hepatocellular carcinoma by regulating the UPF1/Smad7 signalling pathway. J. Cell. Mol. Med. 2019, 23, 6271–6282. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Pestova, T.V.; Hellen, C.U.; Tiedge, H. Translational control by a small RNA: Dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol. Cell Biol. 2008, 28, 3008–3019. [Google Scholar] [CrossRef]
- Hu, G.; Lou, Z.; Gupta, M. The long non-coding RNA GAS5 cooperates with the eukaryotic translation initiation factor 4E to regulate c-Myc translation. PLoS ONE 2014, 9, e107016. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.S.; Sharp, P.A. Emerging roles for natural microRNA sponges. Curr. Biol. CB 2010, 20, R858–R861. [Google Scholar] [CrossRef]
- Kallen, A.N.; Zhou, X.B.; Xu, J.; Qiao, C.; Ma, J.; Yan, L.; Lu, L.; Liu, C.; Yi, J.S.; Zhang, H.; et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell 2013, 52, 101–112. [Google Scholar] [CrossRef]
- Wang, W.-T.; Ye, H.; Wei, P.-P.; Han, B.-W.; He, B.; Chen, Z.-H.; Chen, Y.-Q. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J. Hematol. Oncol. 2016, 9, 117. [Google Scholar] [CrossRef]
- Lee, S.; Kopp, F.; Chang, T.C.; Sataluri, A.; Chen, B.; Sivakumar, S.; Yu, H.; Xie, Y.; Mendell, J.T. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell 2016, 164, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, C.Y.; Zhou, L.Y.; Wang, J.X.; Wang, M.; Zhao, B.; Zhao, W.K.; Xu, S.J.; Fan, L.H.; Zhang, X.J.; et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat. Commun. 2015, 6, 6779. [Google Scholar] [CrossRef]
- Li, H.; Liu, X.; Zhang, L.; Li, X. LncRNA BANCR facilitates vascular smooth muscle cell proliferation and migration through JNK pathway. Oncotarget 2017, 8, 114568–114575. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, X.J.; Ji, Y.X.; Zhang, P.; Deng, K.Q.; Gong, J.; Ren, S.; Wang, X.; Chen, I.; Wang, H.; et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 2016, 22, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Viereck, J.; Kumarswamy, R.; Foinquinos, A.; Xiao, K.; Avramopoulos, P.; Kunz, M.; Dittrich, M.; Maetzig, T.; Zimmer, K.; Remke, J.; et al. Long noncoding RNA Chast promotes cardiac remodeling. Sci. Transl. Med. 2016, 8, 326ra322. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, F.; Zhou, L.Y.; Long, B.; Yuan, S.M.; Wang, Y.; Liu, C.Y.; Sun, T.; Zhang, X.J.; Li, P.F. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ. Res. 2014, 114, 1377–1388. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, E.J.; van Solingen, C.; Scacalossi, K.R.; Ouimet, M.; Afonso, M.S.; Prins, J.; Koelwyn, G.J.; Sharma, M.; Ramkhelawon, B.; Carpenter, S.; et al. The long noncoding RNA CHROME regulates cholesterol homeostasis in primate. Nat. Metab. 2019, 1, 98–110. [Google Scholar] [CrossRef]
- Das, S.; Zhang, E.; Senapati, P.; Amaram, V.; Reddy, M.A.; Stapleton, K.; Leung, A.; Lanting, L.; Wang, M.; Chen, Z.; et al. A Novel Angiotensin II-Induced Long Noncoding RNA Giver Regulates Oxidative Stress, Inflammation, and Proliferation in Vascular Smooth Muscle Cells. Circ. Res. 2018, 123, 1298–1312. [Google Scholar] [CrossRef]
- Liao, B.; Chen, R.; Lin, F.; Mai, A.; Chen, J.; Li, H.; Xu, Z.; Dong, S. Long noncoding RNA HOTTIP promotes endothelial cell proliferation and migration via activation of the Wnt/β-catenin pathway. J. Cell. Biochem. 2018, 119, 2797–2805. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, Z.; Zhang, W.; Wang, Y. Long noncoding RNA LINC00968 promotes endothelial cell proliferation and migration via regulating miR-9-3p expression. J. Cell. Biochem. 2018, 120, 8214–8221. [Google Scholar] [CrossRef]
- Wang, H.; Qin, R.; Cheng, Y. LncRNA-Ang362 Promotes Pulmonary Arterial Hypertension by Regulating miR-221 and miR-222. Shock 2020, 53, 723–729. [Google Scholar] [CrossRef]
- Jin, L.; Lin, X.; Yang, L.; Fan, X.; Wang, W.; Li, S.; Li, J.; Liu, X.; Bao, M.; Cui, X.; et al. AK098656, a Novel Vascular Smooth Muscle Cell-Dominant Long Noncoding RNA, Promotes Hypertension. Hypertension 2018, 71, 262–272. [Google Scholar] [CrossRef]
- Qu, X.; Du, Y.; Shu, Y.; Gao, M.; Sun, F.; Luo, S.; Yang, T.; Zhan, L.; Yuan, Y.; Chu, W.; et al. MIAT Is a Pro-fibrotic Long Non-coding RNA Governing Cardiac Fibrosis in Post-infarct Myocardium. Sci. Rep. 2017, 7, 42657. [Google Scholar] [CrossRef]
- Li, X.; Zhou, J.; Huang, K. Inhibition of the lncRNA Mirt1 Attenuates Acute Myocardial Infarction by Suppressing NF-κB Activation. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2017, 42, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tong, Y.; Zhou, Y.; Han, Z.; Wang, X.; Ding, T.; Qu, Y.; Zhang, Z.; Chang, C.; Zhang, X.; et al. LncRNA KCNQ1OT1 as a miR-26a-5p sponge regulates ATG12-mediated cardiomyocyte autophagy and aggravates myocardial infarction. Int. J. Cardiol. 2021, 338, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hu, Q.; Zhang, B.F.; Liu, X.P.; Yang, S.; Jiang, H. Long noncoding RNA UCA1 inhibits ischaemia/reperfusion injury induced cardiomyocytes apoptosis via suppression of endoplasmic reticulum stress. Genes Genom. 2019, 41, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Long, B.; Li, N.; Xu, X.X.; Li, X.X.; Xu, X.J.; Guo, D.; Zhang, D.; Wu, Z.H.; Zhang, S.Y. Long noncoding RNA FTX regulates cardiomyocyte apoptosis by targeting miR-29b-1-5p and Bcl2l2. Biochem. Biophys. Res. Commun. 2018, 495, 312–318. [Google Scholar] [CrossRef]
- Liu, K.; Liu, C.; Zhang, Z. lncRNA GAS5 acts as a ceRNA for miR-21 in suppressing PDGF-bb-induced proliferation and migration in vascular smooth muscle cells. J. Cell. Biochem. 2019, 120, 15233–15240. [Google Scholar] [CrossRef]
- Gao, L.; Liu, Y.; Guo, S.; Yao, R.; Wu, L.; Xiao, L.; Wang, Z.; Liu, Y.; Zhang, Y. Circulating Long Noncoding RNA HOTAIR is an Essential Mediator of Acute Myocardial Infarction. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2017, 44, 1497–1508. [Google Scholar] [CrossRef]
- Li, C.; Lin, L.; Zhang, L.; Xu, R.; Chen, X.; Ji, J.; Li, Y. Long noncoding RNA p21 enhances autophagy to alleviate endothelial progenitor cells damage and promote endothelial repair in hypertension through SESN2/AMPK/TSC2 pathway. Pharmacol. Res. 2021, 173, 105920. [Google Scholar] [CrossRef]
- Wu, G.; Cai, J.; Han, Y.; Chen, J.; Huang, Z.P.; Chen, C.; Cai, Y.; Huang, H.; Yang, Y.; Liu, Y.; et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation 2014, 130, 1452–1465. [Google Scholar] [CrossRef]
- Han, P.; Li, W.; Lin, C.H.; Yang, J.; Shang, C.; Nuernberg, S.T.; Jin, K.K.; Xu, W.; Lin, C.Y.; Lin, C.J.; et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014, 514, 102–106. [Google Scholar] [CrossRef]
- Hu, Y.-W.; Guo, F.-X.; Xu, Y.-J.; Li, P.; Lu, Z.-F.; McVey, D.G.; Zheng, L.; Wang, Q.; Ye, J.H.; Kang, C.-M.; et al. Long noncoding RNA NEXN-AS1 mitigates atherosclerosis by regulating the actin-binding protein NEXN. J. Clin. Investig. 2019, 129, 1115–1128. [Google Scholar] [CrossRef]
- Boulberdaa, M.; Scott, E.; Ballantyne, M.; Garcia, R.; Descamps, B.; Angelini, G.D.; Brittan, M.; Hunter, A.; McBride, M.; McClure, J.; et al. A Role for the Long Noncoding RNA SENCR in Commitment and Function of Endothelial Cells. Mol. Ther. 2016, 24, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Zeng, T.; Ni, H.; Yu, Y.; Zhang, M.; Wu, M.; Wang, Q.; Wang, L.; Xu, S.; Xu, Z.; Xu, C.; et al. BACE1-AS prevents BACE1 mRNA degradation through the sequestration of BACE1-targeting miRNAs. J. Chem. Neuroanat. 2019, 98, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Yu, X.; Wang, M.; Meng, Y.; Song, D.; Yang, H.; Wang, D.; Bi, J.; Xu, S. Long Non-coding RNAs in Pathogenesis of Neurodegenerative Diseases. Front. Cell Dev. Biol. 2021, 9, 719247. [Google Scholar] [CrossRef] [PubMed]
- Kovanda, A.; Zalar, M.; Šket, P.; Plavec, J.; Rogelj, B. Anti-sense DNA d (GGCCCC) n expansions in C9ORF72 form i-motifs and protonated hairpins. Sci. Rep. 2015, 5, 17944. [Google Scholar] [CrossRef]
- Gu, C.; Chen, C.; Wu, R.; Dong, T.; Hu, X.; Yao, Y.; Zhang, Y. Long Noncoding RNA EBF3-AS Promotes Neuron Apoptosis in Alzheimer’s Disease. DNA Cell Biol. 2018, 37, 220–226. [Google Scholar] [CrossRef]
- Xu, W.; Zhang, L.; Geng, Y.; Liu, Y.; Zhang, N. Long noncoding RNA GAS5 promotes microglial inflammatory response in Parkinson’s disease by regulating NLRP3 pathway through sponging miR-223-3p. Int. Immunopharmacol. 2020, 85, 106614. [Google Scholar] [CrossRef]
- Yan, Y.; Yan, H.; Teng, Y.; Wang, Q.; Yang, P.; Zhang, L.; Cheng, H.; Fu, S. Long non-coding RNA 00507/miRNA-181c-5p/TTBK1/MAPT axis regulates tau hyperphosphorylation in Alzheimer’s disease. J. Gene Med. 2020, 22, e3268. [Google Scholar] [CrossRef]
- Xu, X.; Zhuang, C.; Wu, Z.; Qiu, H.; Feng, H.; Wu, J. LincRNA-p21 Inhibits Cell Viability and Promotes Cell Apoptosis in Parkinson’s Disease through Activating α-Synuclein Expression. BioMed Res. Int. 2018, 2018, 8181374. [Google Scholar] [CrossRef]
- Zhang, Q.S.; Wang, Z.H.; Zhang, J.L.; Duan, Y.L.; Li, G.F.; Zheng, D.L. Beta-asarone protects against MPTP-induced Parkinson’s disease via regulating long non-coding RNA MALAT1 and inhibiting α-synuclein protein expression. Biomed. Pharmacother. 2016, 83, 153–159. [Google Scholar] [CrossRef]
- DeWitt, J.J.; Grepo, N.; Wilkinson, B.; Evgrafov, O.V.; Knowles, J.A.; Campbell, D.B. Impact of the Autism-Associated Long Noncoding RNA MSNP1AS on Neuronal Architecture and Gene Expression in Human Neural Progenitor Cells. Genes 2016, 7, 76. [Google Scholar] [CrossRef]
- Simchovitz, A.; Hanan, M.; Niederhoffer, N.; Madrer, N.; Yayon, N.; Bennett, E.R.; Greenberg, D.S.; Kadener, S.; Soreq, H. NEAT1 is overexpressed in Parkinson’s disease substantia nigra and confers drug-inducible neuroprotection from oxidative stress. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2019, 33, 11223–11234. [Google Scholar] [CrossRef] [PubMed]
- Luo, T.; Liu, P.; Wang, X.Y.; Li, L.Z.; Zhao, L.P.; Huang, J.; Li, Y.M.; Ou, J.L.; Peng, X.Q. Effect of the autism-associated lncRNA Shank2-AS on architecture and growth of neurons. J. Cell. Biochem. 2019, 120, 1754–1762. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lu, B.; Chen, J. Knockdown of lncRNA SNHG1 attenuated Aβ(25-35)-inudced neuronal injury via regulating KREMEN1 by acting as a ceRNA of miR-137 in neuronal cells. Biochem. Biophys. Res. Commun. 2019, 518, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lian, Y.J.; Ma, Y.Q.; Wu, C.J.; Zheng, Y.K.; Xie, N.C. LncRNA SNHG1 promotes α-synuclein aggregation and toxicity by targeting miR-15b-5p to activate SIAH1 in human neuroblastoma SH-SY5Y cells. Neurotoxicology 2018, 68, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Wang, T.; Qu, Q.; Kang, T.; Yang, Q. Long Noncoding RNA SNHG1 Promotes Neuroinflammation in Parkinson’s Disease via Regulating miR-7/NLRP3 Pathway. Neuroscience 2018, 388, 118–127. [Google Scholar] [CrossRef]
- Zhang, L.M.; Wang, M.H.; Yang, H.C.; Tian, T.; Sun, G.F.; Ji, Y.F.; Hu, W.T.; Liu, X.; Wang, J.P.; Lu, H. Dopaminergic neuron injury in Parkinson’s disease is mitigated by interfering lncRNA SNHG14 expression to regulate the miR-133b/α-synuclein pathway. Aging 2019, 11, 9264–9279. [Google Scholar] [CrossRef]
- Xu, W.; Li, K.; Fan, Q.; Zong, B.; Han, L. Knockdown of long non-coding RNA SOX21-AS1 attenuates amyloid-β-induced neuronal damage by sponging miR-107. Biosci. Rep. 2020, 40, BSR20194295. [Google Scholar] [CrossRef]
- Velmeshev, D.; Magistri, M.; Faghihi, M.A. Expression of non-protein-coding antisense RNAs in genomic regions related to autism spectrum disorders. Mol. Autism 2013, 4, 32. [Google Scholar] [CrossRef]
- Yue, D.; Guanqun, G.; Jingxin, L.; Sen, S.; Shuang, L.; Yan, S.; Minxue, Z.; Ping, Y.; Chong, L.; Zhuobo, Z.; et al. Silencing of long noncoding RNA XIST attenuated Alzheimer’s disease-related BACE1 alteration through miR-124. Cell Biol. Int. 2020, 44, 630–636. [Google Scholar] [CrossRef]
- Aliperti, V.; Skonieczna, J.; Cerase, A. Long Non-Coding RNA (lncRNA) Roles in Cell Biology, Neurodevelopment and Neurological Disorders. Noncoding RNA 2021, 7, 36. [Google Scholar] [CrossRef]
- Jiang, J.; Piao, X.; Hu, S.; Gao, J.; Bao, M. LncRNA H19 diminishes dopaminergic neuron loss by mediating microRNA-301b-3p in Parkinson’s disease via the HPRT1-mediated Wnt/β-catenin signaling pathway. Aging 2020, 12, 8820–8836. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Li, Y.; Zhang, W.; Fang, F.; Sun, J.; Liu, M.; Li, K.; Dong, L. Long Non-Coding RNA MALAT1 Inhibits Neuron Apoptosis and Neuroinflammation While Stimulates Neurite Outgrowth and Its Correlation with miR-125b Mediates PTGS2, CDK5 and FOXQ1 in Alzheimer’s Disease. Curr. Alzheimer Res. 2019, 16, 596–612. [Google Scholar] [CrossRef]
- Yi, J.; Chen, B.; Yao, X.; Lei, Y.; Ou, F.; Huang, F. Upregulation of the lncRNA MEG3 improves cognitive impairment, alleviates neuronal damage, and inhibits activation of astrocytes in hippocampus tissues in Alzheimer’s disease through inactivating the PI3K/Akt signaling pathway. J. Cell. Biochem. 2019, 120, 18053–18065. [Google Scholar] [CrossRef] [PubMed]
- Ip, J.Y.; Sone, M.; Nashiki, C.; Pan, Q.; Kitaichi, K.; Yanaka, K.; Abe, T.; Takao, K.; Miyakawa, T.; Blencowe, B.J.; et al. Gomafu lncRNA knockout mice exhibit mild hyperactivity with enhanced responsiveness to the psychostimulant methamphetamine. Sci. Rep. 2016, 6, 27204. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chen, Y.; Huang, Y.; Cao, K.; Liu, T.; Shen, H.; Cui, J.; Li, B.; Cai, J.; Gao, F.; et al. Long non-coding RNA ANRIL promotes homologous recombination-mediated DNA repair by maintaining ATR protein stability to enhance cancer resistance. Mol. Cancer 2021, 20, 94. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Mi, S.; Zhao, T.; Peng, C.; Peng, Y.; Chen, L.; Zhu, W.; Yao, Y.; Song, Q.; Li, X.; et al. BGL3 lncRNA mediates retention of the BRCA1/BARD1 complex at DNA damage sites. EMBO J. 2020, 39, e104133. [Google Scholar] [CrossRef]
- Chen, L.; Wang, W.; Cao, L.; Li, Z.; Wang, X. Long Non-Coding RNA CCAT1 Acts as a Competing Endogenous RNA to Regulate Cell Growth and Differentiation in Acute Myeloid Leukemia. Mol. Cells 2016, 39, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Spizzo, R.; Atlasi, Y.; Nicoloso, M.; Shimizu, M.; Redis, R.S.; Nishida, N.; Gafà, R.; Song, J.; Guo, Z.; et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013, 23, 1446–1461. [Google Scholar] [CrossRef] [PubMed]
- Ellis, B.C.; Graham, L.D.; Molloy, P.L. CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochim. Biophys. Acta BBA Mol. Cell Res. 2014, 1843, 372–386. [Google Scholar] [CrossRef] [PubMed]
- Klein, U.; Lia, M.; Crespo, M.; Siegel, R.; Shen, Q.; Mo, T.; Ambesi-Impiombato, A.; Califano, A.; Migliazza, A.; Bhagat, G.; et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 2010, 17, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Xiao, Z.; Wang, Y.; Zheng, M.; Song, T.; Cai, X.; Sun, B.; Ye, L.; Zhang, X. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015, 75, 846–857. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.; Li, Q.; Fang, J.; Zhao, T. LncRNA HOTAIR: A Potential Prognostic Factor and Therapeutic Target in Human Cancers. Front. Oncol. 2021, 11, 679244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cai, K.; Wang, J.; Wang, X.; Cheng, K.; Shi, F.; Jiang, L.; Zhang, Y.; Dou, J. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells 2014, 32, 2858–2868. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, M.; Groß, M.; Holler, J.M.; Coggins, S.A.; Patil, N.; Leupold, J.H.; Munschauer, M.; Schenone, M.; Hartigan, C.R.; Allgayer, H.; et al. The lncRNA lincNMR regulates nucleotide metabolism via a YBX1—RRM2 axis in cancer. Nat. Commun. 2020, 11, 3214. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Nie, J.; Lu, L.; Du, C.; Meng, F.; Song, D. LINC00337 promotes tumor angiogenesis in colorectal cancer by recruiting DNMT1, which suppresses the expression of CNN1. Cancer Gene Ther. 2021, 28, 1285–1297. [Google Scholar] [CrossRef]
- Zhang, N.; Zeng, X.; Sun, C.; Guo, H.; Wang, T.; Wei, L.; Zhang, Y.; Zhao, J.; Ma, X. LncRNA LINC00963 Promotes Tumorigenesis and Radioresistance in Breast Cancer by Sponging miR-324-3p and Inducing ACK1 Expression. Mol. Ther. Nucleic Acids 2019, 18, 871–881. [Google Scholar] [CrossRef]
- Zhou, M.; Hou, Y.; Yang, G.; Zhang, H.; Tu, G.; Du, Y.E.; Wen, S.; Xu, L.; Tang, X.; Tang, S.; et al. LncRNA-Hh Strengthen Cancer Stem Cells Generation in Twist-Positive Breast Cancer via Activation of Hedgehog Signaling Pathway. Stem Cells 2016, 34, 55–66. [Google Scholar] [CrossRef]
- Chen, C.; He, W.; Huang, J.; Wang, B.; Li, H.; Cai, Q.; Su, F.; Bi, J.; Liu, H.; Zhang, B.; et al. LNMAT1 promotes lymphatic metastasis of bladder cancer via CCL2 dependent macrophage recruitment. Nat. Commun. 2018, 9, 3826. [Google Scholar] [CrossRef]
- Wang, Q.M.; Lian, G.Y.; Song, Y.; Huang, Y.F.; Gong, Y. LncRNA MALAT1 promotes tumorigenesis and immune escape of diffuse large B cell lymphoma by sponging miR-195. Life Sci. 2019, 231, 116335. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Ou, C.; Liu, J.; Chen, C.; Zhou, Q.; Yang, S.; Li, G.; Wang, G.; Song, J.; Li, Z.; et al. YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene 2019, 38, 2627–2644. [Google Scholar] [CrossRef] [PubMed]
- Goyal, B.; Yadav, S.R.M.; Awasthee, N.; Gupta, S.; Kunnumakkara, A.B.; Gupta, S.C. Diagnostic, prognostic, and therapeutic significance of long non-coding RNA MALAT1 in cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188502. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Yan, B.; Lu, Q.; Lin, Y.; Ma, L. The role of MALAT1/miR-1/slug axis on radioresistance in nasopharyngeal carcinoma. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2016, 37, 4025–4033. [Google Scholar] [CrossRef]
- Yuan, S.X.; Yang, F.; Yang, Y.; Tao, Q.F.; Zhang, J.; Huang, G.; Yang, Y.; Wang, R.Y.; Yang, S.; Huo, X.S.; et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology 2012, 56, 2231–2241. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Fu, Y.; Zhu, N.; Wang, Z.; Hong, J.-L.; Li, Y.; Li, W.-J.; Zhang, H.-B.; Song, J.-H. Repression of lncRNA NEAT1 enhances the antitumor activity of CD8+ T cells against hepatocellular carcinoma via regulating miR-155/Tim-3. Int. J. Biochem. Cell Biol. 2019, 110, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Lang, H.L.; Hu, G.W.; Chen, Y.; Liu, Y.; Tu, W.; Lu, Y.M.; Wu, L.; Xu, G.H. Glioma cells promote angiogenesis through the release of exosomes containing long non-coding RNA POU3F3. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 959–972. [Google Scholar] [PubMed]
- Chen, J.; Yu, Y.; Li, H.; Hu, Q.; Chen, X.; He, Y.; Xue, C.; Ren, F.; Ren, Z.; Li, J.; et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol. Cancer 2019, 18, 33. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Du, P.; Cui, P.; Qin, Y.; Wu, J.; Zhou, Z.; Zhang, W.; Qin, L.; Huang, G. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene 2018, 37, 4094–4109. [Google Scholar] [CrossRef] [PubMed]
- Yari, H.; Jin, L.; Teng, L.; Wang, Y.; Wu, Y.; Liu, G.Z.; Gao, W.; Liang, J.; Xi, Y.; Feng, Y.C.; et al. LncRNA REG1CP promotes tumorigenesis through an enhancer complex to recruit FANCJ helicase for REG3A transcription. Nat. Commun. 2019, 10, 5334. [Google Scholar] [CrossRef]
- He, X.; Wang, J.; Chen, J.; Han, L.; Lu, X.; Miao, D.; Yin, D.; Geng, Q.; Zhang, E. lncRNA UCA1 Predicts a Poor Prognosis and Regulates Cell Proliferation and Migration by Repressing p21 and SPRY1 Expression in GC. Mol. Nucleic Acids 2019, 18, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Li, H.J.; Li, X.; Pang, H.; Pan, J.J.; Xie, X.J.; Chen, W. Long non-coding RNA UCA1 promotes glutamine metabolism by targeting miR-16 in human bladder cancer. Jpn. J. Clin. Oncol. 2015, 45, 1055–1063. [Google Scholar] [CrossRef]
- Gao, Z.; Wang, Q.; Ji, M.; Guo, X.; Li, L.; Su, X. Exosomal lncRNA UCA1 modulates cervical cancer stem cell self-renewal and differentiation through microRNA-122-5p/SOX2 axis. J. Transl. Med. 2021, 19, 229. [Google Scholar] [CrossRef]
- Fang, Q.; Chen, X.; Zhi, X. Long Non-Coding RNA (LncRNA) Urothelial Carcinoma Associated 1 (UCA1) Increases Multi-Drug Resistance of Gastric Cancer via Downregulating miR-27b. Med. Sci. Monit. 2016, 22, 3506–3513. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Wu, L.; Wang, L.; Yang, Y.; Meng, Y.; Yang, H. Increased expression of the long non-coding RNA UCA1 in tongue squamous cell carcinomas: A possible correlation with cancer metastasis. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2014, 117, 89–95. [Google Scholar] [CrossRef]
- Li, S.; Zhou, J.; Wang, Z.; Wang, P.; Gao, X.; Wang, Y. Long noncoding RNA GAS5 suppresses triple negative breast cancer progression through inhibition of proliferation and invasion by competitively binding miR-196a-5p. Biomed. Pharmacother. 2018, 104, 451–457. [Google Scholar] [CrossRef]
- Chen, X.; Xie, R.; Gu, P.; Huang, M.; Han, J.; Dong, W.; Xie, W.; Wang, B.; He, W.; Zhong, G.; et al. Long Noncoding RNA LBCS Inhibits Self-Renewal and Chemoresistance of Bladder Cancer Stem Cells through Epigenetic Silencing of SOX2. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 1389–1403. [Google Scholar] [CrossRef]
- Dimitrova, N.; Zamudio, J.R.; Jong, R.M.; Soukup, D.; Resnick, R.; Sarma, K.; Ward, A.J.; Raj, A.; Lee, J.T.; Sharp, P.A.; et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol. Cell 2014, 54, 777–790. [Google Scholar] [CrossRef] [PubMed]
- Ding, G.; Peng, Z.; Shang, J.; Kang, Y.; Ning, H.; Mao, C. LincRNA-p21 inhibits invasion and metastasis of hepatocellular carcinoma through miR-9/E-cadherin cascade signaling pathway molecular mechanism. Onco Targets 2017, 10, 3241–3247. [Google Scholar] [CrossRef] [PubMed]
- Alipoor, B.; Nikouei, S.; Rezaeinejad, F.; Malakooti-Dehkordi, S.N.; Sabati, Z.; Ghasemi, H. Long non-coding RNAs in metabolic disorders: Pathogenetic relevance and potential biomarkers and therapeutic targets. J. Endocrinol. Investig. 2021, 44, 2015–2041. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Zhang, Y.; Zhang, X.; Yao, Y.; Zheng, Y.; Cui, X.; Liu, C.; Wang, Q.; Li, J.Z. Long non-coding RNA Bhmt-AS attenuates hepatic gluconeogenesis via modulation of Bhmt expression. Biochem. Biophys. Res. Commun. 2019, 516, 215–221. [Google Scholar] [CrossRef]
- Li, M.; Guo, Y.; Wang, X.J.; Duan, B.H.; Li, L. HOTAIR participates in hepatic insulin resistance via regulating SIRT1. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7883–7890. [Google Scholar] [CrossRef]
- Zhao, Z.; Liu, B.; Li, B.; Song, C.; Diao, H.; Guo, Z.; Li, Z.; Zhang, J. Inhibition of long noncoding RNA IGF2AS promotes angiogenesis in type 2 diabetes. Biomed. Pharmacother. 2017, 92, 445–450. [Google Scholar] [CrossRef]
- Yang, F.; Qin, Y.; Wang, Y.; Li, A.; Lv, J.; Sun, X.; Che, H.; Han, T.; Meng, S.; Bai, Y.; et al. LncRNA KCNQ1OT1 Mediates Pyroptosis in Diabetic Cardiomyopathy. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 50, 1230–1244. [Google Scholar] [CrossRef]
- Kong, X.; Liu, C.-X.; Wang, G.-D.; Yang, H.; Yao, X.-M.; Hua, Q.; Li, X.-Y.; Zhang, H.-M.; Ma, M.-Z.; Su, Q. LncRNA LEGLTBC functions as a ceRNA to antagonize the effects of miR-34a on the downregulation of SIRT1 in glucolipotoxicity-induced INS-1 beta cell oxidative stress and apoptosis. Oxidative Med. Cell. Longev. 2019, 2019, 4010764. [Google Scholar] [CrossRef]
- Li, Z.; Yu, Z.; Meng, X.; Yu, P. LncRNA LINC00968 accelerates the proliferation and fibrosis of diabetic nephropathy by epigenetically repressing p21 via recruiting EZH2. Biochem. Biophys. Res. Commun. 2018, 504, 499–504. [Google Scholar] [CrossRef]
- Ruan, Y.; Lin, N.; Ma, Q.; Chen, R.; Zhang, Z.; Wen, W.; Chen, H.; Sun, J. Circulating LncRNAs analysis in patients with type 2 diabetes reveals novel genes influencing glucose metabolism and islet β-cell function. Cell. Physiol. Biochem. 2018, 46, 335–350. [Google Scholar] [CrossRef]
- Zhang, P.; Sun, Y.; Peng, R.; Chen, W.; Fu, X.; Zhang, L.; Peng, H.; Zhang, Z. Long non-coding RNA Rpph1 promotes inflammation and proliferation of mesangial cells in diabetic nephropathy via an interaction with Gal-3. Cell Death Dis. 2019, 10, 526. [Google Scholar] [CrossRef]
- Yan, C.; Chen, J.; Chen, N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci. Rep. 2016, 6, 22640. [Google Scholar] [CrossRef]
- Liu, B.; Qiang, L.; Wang, G.; Duan, Q.; Liu, J. LncRNA MALAT1 facilities high glucose induced endothelial to mesenchymal transition and fibrosis via targeting miR-145/ZEB2 axis. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3478–3486. [Google Scholar]
- Liu, P.; Jia, S.-B.; Shi, J.-M.; Li, W.-J.; Tang, L.-S.; Zhu, X.-H.; Tong, P. LncRNA-MALAT1 promotes neovascularization in diabetic retinopathy through regulating miR-125b/VE-cadherin axis. Biosci. Rep. 2019, 39, bsr20181469. [Google Scholar] [CrossRef]
- Han, Y.; Qiu, H.; Pei, X.; Fan, Y.; Tian, H.; Geng, J. Low-Dose Sinapic Acid Abates the Pyroptosis of Macrophages by Downregulation of lncRNA-MALAT1 in Rats with Diabetic Atherosclerosis. J. Cardiovasc. Pharmacol. 2018, 71, 104–112. [Google Scholar] [CrossRef]
- Wang, L.-Q.; Zhou, H.-J. LncRNA MALAT1 promotes high glucose-induced inflammatory response of microglial cells via provoking MyD88/IRAK1/TRAF6 signaling. Sci. Rep. 2018, 8, 8346. [Google Scholar] [CrossRef]
- Zhu, X.; Li, H.; Wu, Y.; Zhou, J.; Yang, G.; Wang, W. lncRNA MEG3 promotes hepatic insulin resistance by serving as a competing endogenous RNA of miR-214 to regulate ATF4 expression. Int. J. Mol. Med. 2019, 43, 345–357. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, Z.; Zhu, D.; Zhao, W.; Li, F. Long non-coding RNA MEG3 serves as a ceRNA for microRNA-145 to induce apoptosis of AC16 cardiomyocytes under high glucose condition. Biosci. Rep. 2019, 39, bsr20190444. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, W.; Jin, M.; Chen, J.; Xu, W.; Kong, X. lncRNA MIAT functions as a competing endogenous RNA to upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy. Cell Death Dis. 2017, 8, e2929. [Google Scholar] [CrossRef]
- Peng, H.; Zou, L.; Xie, J.; Wu, H.; Wu, B.; Zhu, G.; Lv, Q.; Zhang, X.; Liu, S.; Li, G.; et al. lncRNA NONRATT021972 siRNA Decreases Diabetic Neuropathic Pain Mediated by the P2X(3) Receptor in Dorsal Root Ganglia. Mol. Neurobiol. 2017, 54, 511–523. [Google Scholar] [CrossRef]
- Liu, D.-W.; Zhang, J.-H.; Liu, F.-X.; Wang, X.-T.; Pan, S.-K.; Jiang, D.-K.; Zhao, Z.-H.; Liu, Z.-S. Silencing of long noncoding RNA PVT1 inhibits podocyte damage and apoptosis in diabetic nephropathy by upregulating FOXA1. Exp. Mol. Med. 2019, 51, 1–15. [Google Scholar] [CrossRef]
- Shan, K.; Li, C.-P.; Liu, C.; Liu, X.; Yan, B. RNCR3: A regulator of diabetes mellitus-related retinal microvascular dysfunction. Biochem. Biophys. Res. Commun. 2017, 482, 777–783. [Google Scholar] [CrossRef]
- Wang, S.; Xu, H.; Zou, L.; Xie, J.; Wu, H.; Wu, B.; Yi, Z.; Lv, Q.; Zhang, X.; Ying, M.; et al. LncRNA uc.48+ is involved in diabetic neuropathic pain mediated by the P2X3 receptor in the dorsal root ganglia. Purinergic Signal. 2016, 12, 139–148. [Google Scholar] [CrossRef]
- Wu, H.; Wen, F.; Jiang, M.; Liu, Q.; Nie, Y. LncRNA uc.48+ is involved in the diabetic immune and inflammatory responses mediated by P2X7 receptor in RAW264. 7 macrophages. Int. J. Mol. Med. 2018, 42, 1152–1160. [Google Scholar]
- Jin, F.; Wang, N.; Zhu, Y.; You, L.; Wang, L.; De, W.; Tang, W. Downregulation of long noncoding RNA Gas5 affects cell cycle and insulin secretion in mouse pancreatic β cells. Cell. Physiol. Biochem. 2017, 43, 2062–2073. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, S.; Zhu, Y. Long noncoding RNA growth arrest-specific transcript 5 alleviates renal fibrosis in diabetic nephropathy by downregulating matrix metalloproteinase 9 through recruitment of enhancer of zeste homolog 2. FASEB J. 2020, 34, 2703–2714. [Google Scholar] [CrossRef]
- Gui, W.; Zhu, W.F.; Zhu, Y.; Tang, S.; Zheng, F.; Yin, X.; Lin, X.; Li, H. LncRNAH19 improves insulin resistance in skeletal muscle by regulating heterogeneous nuclear ribonucleoprotein A1. Cell Commun. Signal. 2020, 18, 173. [Google Scholar] [CrossRef]
- Geng, T.; Liu, Y.; Xu, Y.; Jiang, Y.; Zhang, N.; Wang, Z.; Carmichael, G.G.; Taylor, H.S.; Li, D.; Huang, Y. H19 lncRNA Promotes Skeletal Muscle Insulin Sensitivity in Part by Targeting AMPK. Diabetes 2018, 67, 2183–2198. [Google Scholar] [CrossRef]
- Goyal, N.; Tiwary, S.; Kesharwani, D.; Datta, M. Long non-coding RNA H19 inhibition promotes hyperglycemia in mice by upregulating hepatic FoxO1 levels and promoting gluconeogenesis. J. Mol. Med. 2019, 97, 115–126. [Google Scholar] [CrossRef]
- Luo, R.; Xiao, F.; Wang, P.; Hu, Y.X. lncRNA H19 sponging miR-93 to regulate inflammation in retinal epithelial cells under hyperglycemia via XBP1s. Inflamm. Res. Off. J. Eur. Histamine Res. Soc. 2020, 69, 255–265. [Google Scholar] [CrossRef]
- Guo, J.R.; Yin, L.; Chen, Y.Q.; Jin, X.J.; Zhou, X.; Zhu, N.N.; Liu, X.Q.; Wei, H.W.; Duan, L.S. Autologous blood transfusion augments impaired wound healing in diabetic mice by enhancing lncRNA H19 expression via the HIF-1α signaling pathway. Cell Commun. Signal. CCS 2018, 16, 84. [Google Scholar] [CrossRef]
- Zhuo, C.; Jiang, R.; Lin, X.; Shao, M. LncRNA H19 inhibits autophagy by epigenetically silencing of DIRAS3 in diabetic cardiomyopathy. Oncotarget 2017, 8, 1429–1437. [Google Scholar] [CrossRef]
- Qi, K.; Zhong, J. LncRNA HOTAIR improves diabetic cardiomyopathy by increasing viability of cardiomyocytes through activation of the PI3K/Akt pathway. Exp. Ther. Med. 2018, 16, 4817–4823. [Google Scholar] [CrossRef]
- Li, P.; Zhang, N.; Ping, F.; Gao, Y.; Cao, L. lncRNA SCAL1 inhibits inducible nitric oxide synthase in lung cells under high-glucose conditions. Exp. Ther. Med. 2019, 18, 1831–1836. [Google Scholar] [CrossRef]
- You, L.; Wang, N.; Yin, D.; Wang, L.; Jin, F.; Zhu, Y.; Yuan, Q.; De, W. Downregulation of Long Noncoding RNA Meg3 Affects Insulin Synthesis and Secretion in Mouse Pancreatic Beta Cells. J. Cell. Physiol. 2016, 231, 852–862. [Google Scholar] [CrossRef]
- Zhang, D.; Qin, H.; Leng, Y.; Li, X.; Zhang, L.; Bai, D.; Meng, Y.; Wang, J. LncRNA MEG3 overexpression inhibits the development of diabetic retinopathy by regulating TGF-β1 and VEGF. Exp. Ther. Med. 2018, 16, 2337–2342. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, H.; Li, D.; Sun, H.; Li, M.; Hu, H. Long noncoding RNA Mirt2 upregulates USP10 expression to suppress hepatic steatosis by sponging miR-34a-5p. Gene 2019, 700, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, W.; Chen, Z.; Chen, J.; Meng, Y.; Feng, B.; Sun, L.; Dou, L.; Li, J.; Cui, Q. Long noncoding RNA lncSHGL recruits hnRNPA1 to suppress hepatic gluconeogenesis and lipogenesis. Diabetes 2018, 67, 581–593. [Google Scholar] [CrossRef]
- Ke, N.; Pi, L.H.; Liu, Q.; Chen, L. Long Noncoding RNA SNHG7 inhibits high glucose-induced human retinal endothelial cells angiogenesis by regulating miR-543/SIRT1 axis. Biochem. Biophys. Res. Commun. 2019, 514, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gu, M.; Ma, Y.; Peng, Y. LncRNA TUG1 reduces inflammation and enhances insulin sensitivity in white adipose tissue by regulating miR-204/SIRT1 axis in obesity mice. Mol. Cell. Biochem. 2020, 475, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.D.; Zhang, E.B.; You, L.H.; Wang, N.; Wang, L.T.; Jin, F.Y.; Zhu, Y.N.; Cao, L.H.; Yuan, Q.X.; De, W.; et al. Downregulation of lncRNA TUG1 affects apoptosis and insulin secretion in mouse pancreatic β cells. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2015, 35, 1892–1904. [Google Scholar] [CrossRef]
- Song, Y.; Miao, C.; Wang, J. LncRNA ZEB1-AS1 inhibits renal fibrosis in diabetic nephropathy by regulating the miR-217/MAFB axis. RSC Adv. 2019, 9, 30389–30397. [Google Scholar] [CrossRef]
- Ounzain, S.; Micheletti, R.; Arnan, C.; Plaisance, I.; Cecchi, D.; Schroen, B.; Reverter, F.; Alexanian, M.; Gonzales, C.; Ng, S.Y.; et al. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell. Cardiol. 2015, 89, 98–112. [Google Scholar] [CrossRef]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Währisch, S.; Beisaw, A.; Macura, K.; Bläss, G.; Kellis, M.; Werber, M.; et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef]
- Correia, C.C.M.; Rodrigues, L.F.; de Avila Pelozin, B.R.; Oliveira, E.M.; Fernandes, T. Long Non-Coding RNAs in Cardiovascular Diseases: Potential Function as Biomarkers and Therapeutic Targets of Exercise Training. Noncoding RNA 2021, 7, 65. [Google Scholar] [CrossRef]
- Kumarswamy, R.; Bauters, C.; Volkmann, I.; Maury, F.; Fetisch, J.; Holzmann, A.; Lemesle, G.; de Groote, P.; Pinet, F.; Thum, T. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ. Res. 2014, 114, 1569–1575. [Google Scholar] [CrossRef]
- Zhang, Z.; Gao, W.; Long, Q.-Q.; Zhang, J.; Li, Y.-F.; Yan, J.-J.; Yang, Z.-J.; Wang, L.-S. Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population. Sci. Rep. 2017, 7, 7491. [Google Scholar] [CrossRef]
- Chalei, V.; Sansom, S.N.; Kong, L.; Lee, S.; Montiel, J.F.; Vance, K.W.; Ponting, C.P. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. eLife 2014, 3, e04530. [Google Scholar] [CrossRef] [PubMed]
- Modarresi, F.; Faghihi, M.A.; Lopez-Toledano, M.A.; Fatemi, R.P.; Magistri, M.; Brothers, S.P.; van der Brug, M.P.; Wahlestedt, C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 2012, 30, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Ryan, P.; Patel, B.; Makwana, V.; Jadhav, H.R.; Kiefel, M.; Davey, A.; Reekie, T.A.; Rudrawar, S.; Kassiou, M. Peptides, peptidomimetics, and carbohydrate–peptide conjugates as amyloidogenic aggregation inhibitors for Alzheimer’s disease. ACS Chem. Neurosci. 2018, 9, 1530–1551. [Google Scholar] [CrossRef] [PubMed]
- Lashley, T.; Schott, J.M.; Weston, P.; Murray, C.E.; Wellington, H.; Keshavan, A.; Foti, S.C.; Foiani, M.; Toombs, J.; Rohrer, J.D. Molecular biomarkers of Alzheimer’s disease: Progress and prospects. Dis. Models Mech. 2018, 11, dmm031781. [Google Scholar] [CrossRef] [PubMed]
- Janda, E.; Lascala, A.; Carresi, C.; Parafati, M.; Aprigliano, S.; Russo, V.; Savoia, C.; Ziviani, E.; Musolino, V.; Morani, F.; et al. Parkinsonian toxin-induced oxidative stress inhibits basal autophagy in astrocytes via NQO2/quinone oxidoreductase 2: Implications for neuroprotection. Autophagy 2015, 11, 1063–1080. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Ye, Y.; Mao, H.; Yao, L.; Sun, X.; Wang, B.; Zhang, H.; Xie, L.; Zhang, H.; Zhang, Y.; et al. Downregulated lncRNA-SNHG1 enhances autophagy and prevents cell death through the miR-221/222 /p27/mTOR pathway in Parkinson’s disease. Exp. Cell Res. 2019, 384, 111614. [Google Scholar] [CrossRef] [PubMed]
- Shelkovnikova, T.A.; Kukharsky, M.S.; An, H.; Dimasi, P.; Alexeeva, S.; Shabir, O.; Heath, P.R.; Buchman, V.L. Protective paraspeckle hyper-assembly downstream of TDP-43 loss of function in amyotrophic lateral sclerosis. Mol. Neurodegener. 2018, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Fouad, Y.A.; Aanei, C. Revisiting the hallmarks of cancer. Am. J. Cancer Res. 2017, 7, 1016–1036. [Google Scholar] [PubMed]
- Nadhan, R.; Dhanasekaran, D.N. Decoding the Oncogenic Signals from the Long Non-Coding RNAs. Onco 2021, 1, 14. [Google Scholar] [CrossRef]
- Vidoni, C.; Vallino, L.; Ferraresi, A.; Secomandi, E.; Salwa, A.; Chinthakindi, M.; Galetto, A.; Dhanasekaran, D.N.; Isidoro, C. Epigenetic control of autophagy in women’s tumors: Role of non-coding RNAs. J. Cancer Metastasis Treat. 2021, 7, 4. [Google Scholar] [CrossRef]
- Vallino, L.; Ferraresi, A.; Vidoni, C.; Secomandi, E.; Esposito, A.; Dhanasekaran, D.N.; Isidoro, C. Modulation of non-coding RNAs by resveratrol in ovarian cancer cells: In Silico analysis and literature review of the anti-cancer pathways involved. J. Tradit. Complement. Med. 2020, 10, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Taniue, K.; Akimitsu, N. The Functions and Unique Features of LncRNAs in Cancer Development and Tumorigenesis. Int. J. Mol. Sci. 2021, 22, 632. [Google Scholar] [CrossRef]
- Shang, Y.; Feng, B.; Zhou, L.; Ren, G.; Zhang, Z.; Fan, X.; Sun, Y.; Luo, G.; Liang, J.; Wu, K.; et al. The miR27b-CCNG1-P53-miR-508-5p axis regulates multidrug resistance of gastric cancer. Oncotarget 2016, 7, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yu, D.; Li, H.; Lv, Y.; Li, S. Long non-coding RNA UCA1 confers tamoxifen resistance in breast cancer endocrinotherapy through regulation of the EZH2/p21 axis and the PI3K/AKT signaling pathway. Int. J. Oncol. 2019, 54, 1033–1042. [Google Scholar] [CrossRef]
- Fang, Z.; Chen, W.; Yuan, Z.; Liu, X.; Jiang, H. LncRNA-MALAT1 contributes to the cisplatin-resistance of lung cancer by upregulating MRP1 and MDR1 via STAT3 activation. Biomed. Pharmacother. 2018, 101, 536–542. [Google Scholar] [CrossRef]
- Sakran, N.; Graham, Y.; Pintar, T.; Yang, W.; Kassir, R.; Willigendael, E.M.; Singhal, R.; Kooreman, Z.E.; Ramnarain, D.; Mahawar, K.; et al. The many faces of diabetes. Is there a need for re-classification? A narrative review. BMC Endocr. Disord. 2022, 22, 9. [Google Scholar] [CrossRef] [PubMed]
- Ilonen, J.; Lempainen, J.; Veijola, R. The heterogeneous pathogenesis of type 1 diabetes mellitus. Nat. Rev. Endocrinol. 2019, 15, 635–650. [Google Scholar] [CrossRef] [PubMed]
- Ling, C.; Bacos, K.; Ronn, T. Epigenetics of type 2 diabetes mellitus and weight change—A tool for precision medicine? Nat. Rev. Endocrinol. 2022, 18, 433–448. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhong, Q.; Yang, T.; Xiao, X. Improved therapeutic effects on diabetic foot by human mesenchymal stem cells expressing MALAT1 as a sponge for microRNA-205-5p. Aging 2019, 11, 12236. [Google Scholar] [CrossRef] [PubMed]
- Esposito, A.; Ferraresi, A.; Salwa, A.; Vidoni, C.; Dhanasekaran, D.N.; Isidoro, C. Resveratrol Contrasts IL-6 Pro-Growth Effects and Promotes Autophagy-Mediated Cancer Cell Dormancy in 3D Ovarian Cancer: Role of miR-1305 and of Its Target ARH-I. Cancers 2022, 14, 2142. [Google Scholar] [CrossRef] [PubMed]







| Disease | Upregulated LncRNAs | Downregulated LncRNAs |
|---|---|---|
| Cardiovascular Diseases | APF [80] BANCR [81] CHAER [82] CHAST [83] CHRF [84] CHROME [85] Giver [86] HOTTIP [87] LINC00968 [88] lnc-Ang362 [89] lnc-AK098656 [90] MIAT [91] MIRT1 [92] KCNQ1OT1 [93] UCA1 [94] | FTX [95] GAS5 [96] HOTAIR [97] lincRNA-p21 [98,99] MHRT [100] NEXN-AS1 [101] SENCR [102] |
| Neurological Diseases | BACE1-AS [103,104] C9ORF72-AS [105] EBF3-AS [106] GAS5 [107] LINC00507 [108] lincRNA-p21 [109] MALAT1 [110] MEG3 [104] MSNP1-AS [111] NEAT1 [104,112] SHANK2-AS [113] SNHG1 [114,115,116] SNHG14 [117] SOX21-AS1 [118] SYNGAP1-AS [119] TUG1 [104] XIST [104,120] | DISC1-AS [121] H19 [122] HAR1 [104] MALAT1 [123] MEG3 [124] MIAT/GOMAFU [125] |
| Cancer | ANRIL [126] BGL3 [127] CCAT1 [128] CCAT2 [129] CRNDE [130] DLEU2 [131] HULC [132] HOTAIR [133,134] linc-NMR [135] LINC00337 [136] LINC00963 [137] lnc-Hh [138] LNMAT1 [139] MALAT1 [140,141,142,143] MVIH [144] NEAT1 [145] POU3F3 [146] PVT1 [147,148] REG1CP [149] UCA1 [150,151,152,153,154] | GAS5 [155] LBCS [156] lincRNA-p21 [157,158] |
| Diabetes | ANRIL [159] ARAP1-AS2 [159] Bhmt-AS [160] CDKN2B-AS1 [159] HOTAIR [159,161] IGF2-AS [162] KCNQ1OT1 [163] LEGLTBC [164] LINC00968 [165] lnc-p3134 [166] lnc-Rpph1 [167] MALAT1 [168,169,170,171,172] MEG3 [173,174] MIAT [175] NONRATT021972 [176] NR-033515 [159] PVT1 [177] RNCR3 [178] uc.48+ [179,180] | ARAP1-AS1 [159] BANCR [159] CASC2 [159] GAS5 [181,182] H19 [183,184,185,186,187,188] HOTAIR [189] LUCAT1/SCAL1 [190] MEG3 [191,192] MIRT2 [193] SHGL [194] SNHG7 [195] SRA [159] TUG1 [196,197] ZEB1-AS1 [198] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nadhan, R.; Isidoro, C.; Song, Y.S.; Dhanasekaran, D.N. Signaling by LncRNAs: Structure, Cellular Homeostasis, and Disease Pathology. Cells 2022, 11, 2517. https://doi.org/10.3390/cells11162517
Nadhan R, Isidoro C, Song YS, Dhanasekaran DN. Signaling by LncRNAs: Structure, Cellular Homeostasis, and Disease Pathology. Cells. 2022; 11(16):2517. https://doi.org/10.3390/cells11162517
Chicago/Turabian StyleNadhan, Revathy, Ciro Isidoro, Yong Sang Song, and Danny N. Dhanasekaran. 2022. "Signaling by LncRNAs: Structure, Cellular Homeostasis, and Disease Pathology" Cells 11, no. 16: 2517. https://doi.org/10.3390/cells11162517