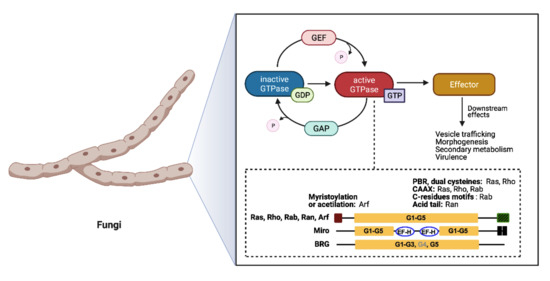
The Small GTPases in Fungal Signaling Conservation and Function
Abstract
:1. Overview
2. Classification and Phylogenetic Conservation of Small Ras GTPases in Fungi
3. Structure of Small Ras GTPases in Fungi
3.1. Ras
3.2. Rho
3.3. Rab
3.4. Ran
3.5. Arf
3.6. Miro
3.7. BRG
4. Small Ras GTPases as Molecular Switches with Multiple Functions in fungi
4.1. Vesicle Trafficking
4.2. Morphogenesis
4.3. Secondary Metabolism
4.4. Virulence
5. The Role of Small GTPases in Fungal-Plant Interactions
5.1. Pathogenic Relationships
5.2. Mutualistic Relationships
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schmoll, M.; Dattenböck, C.; Carreras-Villaseñor, N.; Mendoza-Mendoza, A.; Tisch, D.; Alemán, M.I.; Baker, S.E.; Brown, C.; Cervantes-Badillo, M.G.; Cetz-Chel, J.; et al. The genomes of three uneven siblings: Footprints of the lifestyles of three Trichoderma species. Microbiol. Mol. Biol. Rev. 2016, 80, 205–327. [Google Scholar] [CrossRef] [Green Version]
- Barrett, T.; Xiao, B.; Dodson, E.J.; Dodson, G.; Ludbrook, S.B.; Nurmahomed, K.; Gamblin, S.J.; Musacchio, A.; Smerdon, S.J.; Eccleston, J.F. The structure of the GTPase-activating domain from p50rhoGAP. Nature 1997, 385, 458. [Google Scholar] [CrossRef]
- Wennerberg, K.; Rossman, K.L.; Der, C.J. The Ras superfamily at a glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Dam, T.J.P.; Bos, J.; Snel, B. Evolution of the Ras-like small GTPases and their regulators. Small GTPases 2011, 2, 4–16. [Google Scholar] [CrossRef] [Green Version]
- Francis, S.M.; Gas, M.-E.; Daugeron, M.-C.; Bravo, J.; Seraphin, B. Rbg1–Tma46 dimer structure reveals new functional domains and their role in polysome recruitment. Nucleic Acids Res. 2012, 40, 11100–11114. [Google Scholar] [CrossRef] [Green Version]
- Bourne, H.R.; Sanders, D.A.; McCormick, F. The GTPase superfamily: Conserved structure and molecular mechanism. Nature 1991, 349, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Macara, I.G.; Lounsbury, K.M.; Richards, S.A.; McKiernan, C.; Bar-Sagi, D. The Ras superfamily of GTPases 1. FASEB J. 1996, 10, 625–630. [Google Scholar] [CrossRef] [Green Version]
- Wittinghofer, A.; Vetter, I.R. Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 2011, 80, 943–971. [Google Scholar] [CrossRef]
- Just, W.W.; Peränen, J. Small GTPases in peroxisome dynamics. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2016, 1863, 1006–1013. [Google Scholar] [CrossRef]
- Pereira-Leal, J.B.; Seabra, M.C. Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 2001, 313, 889–901. [Google Scholar] [CrossRef] [Green Version]
- Rojas, A.M.; Fuentes, G.; Rausell, A.; Valencia, A. The Ras protein superfamily: Evolutionary tree and role of conserved amino acids. J. Cell Biol. 2012, 196, 189–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fransson, Å.; Ruusala, A.; Aspenström, P. Atypical Rho GTPases have roles in mitochondrial homeostasis and apoptosis. J. Biol. Chem. 2003, 278, 6495–6502. [Google Scholar] [CrossRef] [Green Version]
- Takai, Y.; Sasaki, T.; Matozaki, T. Small GTP-binding proteins. Physiol. Rev. 2001, 81, 153–208. [Google Scholar] [CrossRef]
- Schmidt, S.; Sohrmann, M.; Hofmann, K.; Woollard, A.; Simanis, V. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 1997, 11, 1519–1534. [Google Scholar] [CrossRef] [Green Version]
- Dautt-Castro, M.; Estrada-Rivera, M.; Olguin-Martínez, I.; del Rocha-Medina, M.C.; Islas-Osuna, M.A.; Casas-Flores, S. TBRG-1 a Ras-like protein in Trichoderma virens involved in conidiation, development, secondary metabolism, mycoparasitism, and biocontrol unveils a new family of Ras-GTPases. Fungal Genet. Biol. 2020, 136, 103292. [Google Scholar] [CrossRef]
- Berbee, M.L.; James, T.Y.; Strullu-Derrien, C. Early diverging fungi: Diversity and impact at the dawn of terrestrial life. Annu. Rev. Microbiol. 2017, 71, 41–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, T.Y.; Stajich, J.E.; Hittinger, C.T.; Rokas, A. Toward a fully resolved fungal tree of life. Annu. Rev. Microbiol. 2020, 74. [Google Scholar] [CrossRef]
- Richards, T.A.; Talbot, N.J. Osmotrophy. Curr. Biol. 2018, 28, R1179–R1180. [Google Scholar] [CrossRef] [Green Version]
- Rokas, A.; Mead, M.E.; Steenwyk, J.L.; Raja, H.A.; Oberlies, N.H. Biosynthetic gene clusters and the evolution of fungal chemodiversity. Nat. Prod. Rep. 2020, 37, 868–878. [Google Scholar] [CrossRef]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014, 42, D699–D704. [Google Scholar] [CrossRef]
- Vernoud, V.; Horton, A.C.; Yang, Z.; Nielsen, E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 2003, 131, 1191–1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores, A.C.; Via, V.D.; Savy, V.; Villagra, U.M.; Zanetti, M.E.; Blanco, F. Comparative phylogenetic and expression analysis of small GTPases families in legume and non-legume plants. Plant Signal. Behav. 2018, 13, e1432956. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.-Y.; Ramachandran, S. Comparative and evolutionary analysis of genes encoding small GTPases and their activating proteins in eukaryotic genomes. Physiol. Genom. 2006, 24, 235–251. [Google Scholar] [CrossRef] [Green Version]
- Kelkar, Y.D.; Ochman, H. Causes and consequences of genome expansion in fungi. Genome Biol. Evol. 2012, 4, 13–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajashekar, B.; Kohler, A.; Johansson, T.; Martin, F.; Tunlid, A.; Ahrén, D. Expansion of signal pathways in the ectomycorrhizal fungus Laccaria bicolor—evolution of nucleotide sequences and expression patterns in families of protein kinases and RAS small GTPases. New Phytol. 2009, 183, 365–379. [Google Scholar] [CrossRef]
- Katinka, M.D.; Duprat, S.; Cornillot, E.; Méténier, G.; Thomarat, F.; Prensier, G.; Barbe, V.; Peyretaillade, E.; Brottier, P.; Wincker, P. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 2001, 414, 450–453. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Repasky, G.A.; Chenette, E.J.; Der, C.J. Renewing the conspiracy theory debate: Does Raf function alone to mediate Ras oncogenesis? Trends Cell Biol. 2004, 14, 639–647. [Google Scholar] [CrossRef]
- Bourne, H.R.; Sanders, D.A.; McCormick, F. The GTPase superfamily: A conserved switch for diverse cell functions. Nature 1990, 348, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Reuther, G.W.; Der, C.J. The Ras branch of small GTPases: Ras family members don’t fall far from the tree. Curr. Opin. Cell Biol. 2000, 12, 157–165. [Google Scholar] [CrossRef]
- Hancock, J.F.; Paterson, H.; Marshall, C.J. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 1990, 63, 133–139. [Google Scholar] [CrossRef]
- Hancock, J.F.; Magee, A.I.; Childs, J.E.; Marshall, C.J. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell 1989, 57, 1167–1177. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Weeks, G.; Spiegelman, G.B. Roles played by Ras subfamily proteins in the cell and developmental biology of microorganisms. Cell. Signal. 2003, 15, 901–909. [Google Scholar] [CrossRef]
- Madaule, P.; Axel, R.; Myers, A.M. Characterization of two members of the rho gene family from the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1987, 84, 779–783. [Google Scholar] [CrossRef] [Green Version]
- Park, H.-O.; Bi, E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 2007, 71, 48–96. [Google Scholar] [CrossRef] [Green Version]
- Harris, S.D. Cdc42/Rho GTPases in fungi: Variations on a common theme. Mol. Microbiol. 2011, 79, 1123–1127. [Google Scholar] [CrossRef]
- Eliás, M.; Klimes, V. Rho GTPases: Deciphering the Evolutionary History of a Complex Protein Family. In Rho GTPases; Springer: New York, NY, USA, 2012; Volume 827, pp. 13–24. [Google Scholar] [CrossRef]
- Zerial, M.; McBride, H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001, 2, 107. [Google Scholar] [CrossRef]
- Pereira-Leal, J.B. The Ypt/Rab family and the evolution of trafficking in fungi. Traffic 2008, 9, 27–38. [Google Scholar] [CrossRef]
- Quimby, B.B.; Dasso, M. The small GTPase Ran: Interpreting the signs. Curr. Opin. Cell Biol. 2003, 15, 338–344. [Google Scholar] [CrossRef]
- Dasso, M. The Ran GTPase: Theme and variations. Curr. Biol. 2002, 12, R502–R508. [Google Scholar] [CrossRef] [Green Version]
- Lounsbury, K.M.; Richards, S.A.; Carey, K.L.; Macara, I.G. Mutations within the Ran/TC4 GTPase: Effects on regulatory factor interactions and subcellular localization. J. Biol. Chem. 1996, 271, 32834–32841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faini, M.; Beck, R.; Wieland, F.T.; Briggs, J.A.G. Vesicle coats: Structure, function, and general principles of assembly. Trends Cell Biol. 2013, 23, 279–288. [Google Scholar] [CrossRef]
- Donaldson, J.G.; Jackson, C.L. ARF family G proteins and their regulators: Roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 2011, 12, 362–375. [Google Scholar] [CrossRef] [PubMed]
- Nakano, A.; Muramatsu, M. A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J. Cell Biol. 1989, 109, 2677–2691. [Google Scholar] [CrossRef] [Green Version]
- Gillingham, A.K.; Munro, S. The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 2007, 23, 579–611. [Google Scholar] [CrossRef]
- Mishra, A.K.; Lambright, D.G. Invited review: Small GTPases and their GAPs. Biopolymers 2016, 105, 431–448. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.C.S.; Orci, L.; Hamamoto, S.; Futai, E.; Ravazzola, M.; Schekman, R. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell 2005, 122, 605–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zinsmaier, K.E. Mitochondrial Miro GTPases coordinate mitochondrial and peroxisomal dynamics. Small GTPases 2020, 1–27. [Google Scholar] [CrossRef]
- Frederick, R.L.; McCaffery, J.M.; Cunningham, K.W.; Okamoto, K.; Shaw, J.M. Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology via a novel pathway. J. Cell Biol. 2004, 167, 87–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klosowiak, J.L.; Focia, P.J.; Chakravarthy, S.; Landahl, E.C.; Freymann, D.M.; Rice, S.E. Structural coupling of the EF hand and C-terminal GTPase domains in the mitochondrial protein Miro. EMBO Rep. 2013, 14, 968–974. [Google Scholar] [CrossRef]
- Tedersoo, L.; Sánchez-Ramírez, S.; Koljalg, U.; Bahram, M.; Döring, M.; Schigel, D.; May, T.; Ryberg, M.; Abarenkov, K. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Divers. 2018, 90, 135–159. [Google Scholar] [CrossRef] [Green Version]
- Schoch, C.L.; Sung, G.-H.; López-Giráldez, F.; Townsend, J.P.; Miadlikowska, J.; Hofstetter, V.; Robbertse, B.; Matheny, P.B.; Kauff, F.; Wang, Z. The Ascomycota tree of life: A phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst. Biol. 2009, 58, 224–239. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [Green Version]
- Binder, J.X.; Pletscher-Frankild, S.; Tsafou, K.; Stolte, C.; O’Donoghue, S.I.; Schneider, R.; Jensen, L.J. COMPARTMENTS: Unification and visualization of protein subcellular localization evidence. Database 2014, 2014, bau012. [Google Scholar] [CrossRef] [Green Version]
- Higuchi, Y. Membrane traffic related to endosome dynamics and protein secretion in filamentous fungi. Biosci. Biotechnol. Biochem. 2021. [Google Scholar] [CrossRef]
- Segev, N.; Mulholland, J.; Botstein, D. The yeast GTP-binding YPT1 protein and a mammalian counterpart are associated with the secretion machinery. Cell 1988, 52, 915–924. [Google Scholar] [CrossRef]
- Nishikawa, S.; Nakano, A. The GTP-binding Sar1 protein is localized to the early compartment of the yeast secretory pathway. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 1991, 1093, 135–143. [Google Scholar] [CrossRef]
- Fiedler, M.R.M.; Cairns, T.C.; Koch, O.; Kubisch, C.; Meyer, V. Conditional Expression of the Small GTPase ArfA Impacts Secretion, Morphology, Growth, and Actin Ring Position in Aspergillus niger. Front. Microbiol. 2018, 9, 878. [Google Scholar] [CrossRef]
- Marešová, L.; Vydarený, T.; Sychrová, H. Comparison of the influence of small GTPases Arl1 and Ypt6 on yeast cells’ tolerance to various stress factors. FEMS Yeast Res. 2012, 12, 332–340. [Google Scholar] [CrossRef] [Green Version]
- Wakade, R.; Labbaoui, H.; Stalder, D.; Arkowitz, R.A.; Bassilana, M. Overexpression of YPT6 restores invasive filamentous growth and secretory vesicle clustering in a Candida albicans arl1 mutant. Small GTPases 2020, 11, 204–210. [Google Scholar] [CrossRef]
- Lichius, A.; Goryachev, A.B.; Fricker, M.D.; Obara, B.; Castro-Longoria, E.; Read, N.D. CDC-42 and RAC-1 regulate opposite chemotropisms in Neurospora crassa. J. Cell Sci. 2014, 127, 1953–1965. [Google Scholar] [CrossRef] [Green Version]
- Virag, A.; Lee, M.P.; Si, H.; Harris, S.D. Regulation of hyphal morphogenesis by cdc42 and rac1 homologues in Aspergillus nidulans. Mol. Microbiol. 2007, 66, 1579–1596. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, A.S.; Edwards, J.E.; Toenjes, K.A.; Johnson, D.I. Cdc42p GTPase regulates the budded-to-hyphal-form transition and expression of hypha-specific transcripts in Candida albicans. Eukaryot. Cell 2004, 3, 724–734. [Google Scholar] [CrossRef] [Green Version]
- Silva, P.M.; Puerner, C.; Seminara, A.; Bassilana, M.; Arkowitz, R.A. Secretory vesicle clustering in fungal filamentous cells does not require directional growth. Cell Rep. 2019, 28, 2231–2245. [Google Scholar] [CrossRef] [Green Version]
- Wedlich-Soldner, R.; Li, R. Yeast and fungal morphogenesis from an evolutionary perspective. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2008; Volume 19, pp. 224–233. [Google Scholar]
- Sudbery, P.; Gow, N.; Berman, J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004, 12, 317–324. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Perlin, M.H.; Garcia-Pedrajas, M.; Covert, S.F.; Gold, S.E. Genetics of morphogenesis and pathogenic development of Ustilago maydis. Adv. Genet. 2007, 57, 1–47. [Google Scholar] [CrossRef]
- Fortwendel, J.R. Orchestration of morphogenesis in filamentous fungi: Conserved roles for Ras signaling networks. Fungal Biol. Rev. 2015, 29, 54–62. [Google Scholar] [CrossRef] [Green Version]
- Tamanoi, F. Ras signaling in yeast. Genes Cancer 2011, 2, 210–215. [Google Scholar] [CrossRef]
- Inglis, D.O.; Sherlock, G. Ras signaling gets fine-tuned: Regulation of multiple pathogenic traits of Candida albicans. Eukaryot. Cell 2013, 12, 1316–1325. [Google Scholar] [CrossRef] [Green Version]
- Ballou, E.R.; Nichols, C.B.; Miglia, K.J.; Kozubowski, L.; Alspaugh, J.A. Two CDC42 paralogues modulate Cryptococcus neoformans thermotolerance and morphogenesis under host physiological conditions. Mol. Microbiol. 2010, 75, 763–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallim, M.A.; Nichols, C.B.; Fernandes, L.; Cramer, K.L.; Alspaugh, J.A. A Rac homolog functions downstream of Ras1 to control hyphal differentiation and high-temperature growth in the pathogenic fungus Cryptococcus neoformans. Eukaryot. Cell 2005, 4, 1066–1078. [Google Scholar] [CrossRef] [Green Version]
- Som, T.; Kolaparthi, V.S. Developmental decisions in Aspergillus nidulans are modulated by Ras activity. Mol. Cell. Biol. 1994, 14, 5333–5348. [Google Scholar] [CrossRef] [PubMed]
- Osherov, N.; May, G. Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics 2000, 155, 647–656. [Google Scholar]
- Liu, Y.; Bell-Pedersen, D. Circadian rhythms in Neurospora crassa and other filamentous fungi. Eukaryot. Cell 2006, 5, 1184–1193. [Google Scholar] [CrossRef] [Green Version]
- Belden, W.J.; Larrondo, L.F.; Froehlich, A.C.; Shi, M.; Chen, C.-H.; Loros, J.J.; Dunlap, J.C. The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output. Genes Dev. 2007, 21, 1494–1505. [Google Scholar] [CrossRef] [Green Version]
- Kana-Uchi, A.; Yamashiro, C.T.; Tanabe, S.; Murayama, T. A ras homologue of Neurospora crassa regulates morphology. Mol. Gen. Genet. MGG 1997, 254, 427–432. [Google Scholar] [CrossRef]
- Rasmussen, C.G.; Glass, N.L. A Rho-type GTPase, rho-4, is required for septation in Neurospora crassa. Eukaryot. Cell 2005, 4, 1913–1925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasmussen, C.G.; Glass, N.L. Localization of RHO-4 indicates differential regulation of conidial versus vegetative septation in the filamentous fungus Neurospora crassa. Eukaryot. Cell 2007, 6, 1097–1107. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, C.G.; Morgenstein, R.M.; Peck, S.; Glass, N.L. Lack of the GTPase RHO-4 in Neurospora crassa causes a reduction in numbers and aberrant stabilization of microtubules at hyphal tips. Fungal Genet. Biol. 2008, 45, 1027–1039. [Google Scholar] [CrossRef]
- Panepinto, J.C.; Oliver, B.G.; Fortwendel, J.R.; Smith, D.L.H.; Askew, D.S.; Rhodes, J.C. Deletion of the Aspergillus fumigatus gene encoding the Ras-related protein RhbA reduces virulence in a model of invasive pulmonary aspergillosis. Infect. Immun. 2003, 71, 2819–2826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortwendel, J.R.; Juvvadi, P.R.; Rogg, L.E.; Asfaw, Y.G.; Burns, K.A.; Randell, S.H.; Steinbach, W.J. Plasma membrane localization is required for RasA-mediated polarized morphogenesis and virulence of Aspergillus fumigatus. Eukaryot. Cell 2012, 11, 966–977. [Google Scholar] [CrossRef] [Green Version]
- Norton, T.S.; Fortwendel, J.R. Control of Ras-mediated signaling in Aspergillus fumigatus. Mycopathologia 2014, 178, 325–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashby, S.F.; Nowell, W. The fungi of stigmatomycosis. Ann. Bot. 1926, 40, 69–83. [Google Scholar] [CrossRef]
- Wendland, J.; Philippsen, P. Cell polarity and hyphal morphogenesis are controlled by multiple rho-protein modules in the filamentous ascomycete Ashbya gossypii. Genetics 2001, 157, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Köhli, M.; Buck, S.; Schmitz, H.P. The function of two closely related Rho proteins is determined by an atypical switch I region. J. Cell Sci. 2008, 121, 1065–1075. [Google Scholar] [CrossRef] [Green Version]
- Nordmann, D.; Lickfeld, M.; Warnsmann, V.; Wiechert, J.; Jendretzki, A.; Schmitz, H.P. The Small GTP-Binding Proteins Ag Rho2 and Ag Rho5 Regulate Tip-Branching, Maintenance of the Growth Axis and Actin-Ring-Integrity in the Filamentous Fungus Ashbya gossypii. PLoS ONE 2014, 9, e106236. [Google Scholar] [CrossRef] [Green Version]
- Bauer, Y.; Knechtle, P.; Wendland, J.; Helfer, H.; Philippsen, P. A Ras-like GTPase is involved in hyphal growth guidance in the filamentous fungus Ashbya gossypii. Mol. Biol. Cell 2004, 15, 4622–4632. [Google Scholar] [CrossRef] [Green Version]
- Casale, W.L.; Mcconnell, D.G.; Wang, S.Y.; Lee, Y.J.; Linz, J.E. Expression of a gene family in the dimorphic fungus Mucor racemosus which exhibits striking similarity to human ras genes. Mol. Cell. Biol. 1990, 10, 6654–6663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roze, L.V.; Mahanti, N.; Mehigh, R.; McConnell, D.G.; Linz, J.E. Evidence that MRas1 and MRas3 proteins are associated with distinct cellular functions during growth and morphogenesis in the fungus Mucor racemosus. Fungal Genet. Biol. 1999, 28, 171–189. [Google Scholar] [CrossRef]
- Boyce, K.J.; Hynes, M.J.; Andrianopoulos, A. The Ras and Rho GTPases genetically interact to co-ordinately regulate cell polarity during development in Penicillium marneffei. Mol. Microbiol. 2005, 55, 1487–1501. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, L.; Paes, H.C.; Tavares, A.H.; Silva, S.S.; Dantas, A.; Soares, C.M.A.; Torres, F.A.G.; Felipe, M.S.S. Transcriptional profile of ras1 and ras2 and the potential role of farnesylation in the dimorphism of the human pathogen Paracoccidioides brasiliensis. FEMS Yeast Res. 2008, 8, 300–310. [Google Scholar] [CrossRef] [Green Version]
- Haniu, A.E.C.J.; Maricato, J.T.; Mathias, P.P.M.; Castilho, D.G.; Miguel, R.B.; Monteiro, H.P.; Puccia, R.; Batista, W.L. Low concentrations of hydrogen peroxide or nitrite induced of Paracoccidioides brasiliensis cell proliferation in a Ras-dependent manner. PLoS ONE 2013, 8, e69590. [Google Scholar] [CrossRef] [Green Version]
- Truesdell, G.M.; Jones, C.; Holt, T.; Henderson, G.; Dickman, M.B. A Ras protein from a phytopathogenic fungus causes defects in hyphal growth polarity, and induces tumors in mice. Mol. Gen. Genet. MGG 1999, 262, 46–54. [Google Scholar] [CrossRef]
- Chen, C.; Dickman, M.B. Dominant active Rac and dominant negative Rac revert the dominant active Ras phenotype in Colletotrichum trifolii by distinct signalling pathways. Mol. Microbiol. 2004, 51, 1493–1507. [Google Scholar] [CrossRef] [PubMed]
- Ha, Y.; Memmott, S.D.; Dickman, M.B. Functional analysis of Ras in Colletotrichum trifolii. FEMS Microbiol. Lett. 2003, 226, 315–321. [Google Scholar] [CrossRef]
- Chen, C.; Ha, Y.-S.; Min, J.-Y.; Memmott, S.D.; Dickman, M.B. Cdc42 is required for proper growth and development in the fungal pathogen Colletotrichum trifolii. Eukaryot. Cell 2006, 5, 155–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bluhm, B.H.; Zhao, X.; Flaherty, J.E.; Xu, J.R.; Dunkle, L.D. RAS2 regulates growth and pathogenesis in Fusarium graminearum. Mol. Plant. Microbe Interact. 2007, 20, 627–636. [Google Scholar] [CrossRef] [Green Version]
- Park, G.; Xue, C.; Zhao, X.; Kim, Y.; Orbach, M.; Xu, J.-R. Multiple upstream signals converge on the adaptor protein Mst50 in Magnaporthe grisea. Plant Cell 2006, 18, 2822–2835. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Kim, Y.; Park, G.; Xu, J.-R. A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell 2005, 17, 1317–1329. [Google Scholar] [CrossRef] [Green Version]
- Dub, A.M.; Kokkelink, L.; Tudzynski, B.; Tudzynski, P.; Sharon, A. Involvement of Botrytis cinerea small GTPases BcRAS1 and BcRAC in differentiation, virulence, and the cell cycle. Eukaryot. Cell 2013, 12, 1609–1618. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.-R.; Hao, Z.-M.; Wang, L.-H.; Shen, S.; Cao, Z.-Y.; Xin, Y.-Y.; Hou, M.-L.; Gu, S.-Q.; Han, J.-M.; Dong, J.-G. StRas2 regulates morphogenesis, conidiation and appressorium development in Setosphaeria turcica. Microbiol. Res. 2012, 167, 478–486. [Google Scholar] [CrossRef]
- Xie, X.-Q.; Guan, Y.; Ying, S.-H.; Feng, M.-G. Differentiated functions of Ras1 and Ras2 proteins in regulating the germination, growth, conidiation, multi-stress tolerance and virulence of Beauveria bassiana. Environ. Microbiol. 2013, 15, 447–462. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Y.; Zhong, Y.; Qu, Y.; Wang, T. Ras GTPases modulate morphogenesis, sporulation and cellulase gene expression in the cellulolytic fungus Trichoderma reesei. PLoS ONE 2012, 7, e48786. [Google Scholar] [CrossRef]
- Knabe, N.; Jung, E.-M.; Freihorst, D.; Hennicke, F.; Horton, J.S.; Kothe, E. A central role for Ras1 in morphogenesis of the basidiomycete Schizophyllum commune. Eukaryot. Cell 2013, 12, 941–952. [Google Scholar] [CrossRef] [Green Version]
- Schubert, D.; Raudaskoski, M.; Knabe, N.; Kothe, E. Ras GTPase-activating protein gap1 of the homobasidiomycete Schizophyllum commune regulates hyphal growth orientation and sexual development. Eukaryot. Cell 2006, 5, 683–695. [Google Scholar] [CrossRef] [Green Version]
- Takemoto, D.; Tanaka, A.; Scott, B. A p67Phox-like regulator is recruited to control hyphal branching in a fungal-grass mutualistic symbiosis. Plant Cell 2006, 18, 2807–2821. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, A.; Takemoto, D.; Hyon, G.; Park, P.; Scott, B. NoxA activation by the small GTPase RacA is required to maintain a mutualistic symbiotic association between Epichloë festucae and perennial ryegrass. Mol. Microbiol. 2008, 68, 1165–1178. [Google Scholar] [CrossRef]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef]
- Bonifacino, J.S.; Glick, B.S. The mechanisms of vesicle budding and fusion. Cell 2004, 116, 153–166. [Google Scholar] [CrossRef] [Green Version]
- Whyte, J.R.C.; Munro, S. Vesicle tethering complexes in membrane traffic. J. Cell Sci. 2002, 115, 2627–2637. [Google Scholar] [CrossRef] [PubMed]
- Wichmann, H.; Hengst, L.; Gallwitz, D. Endocytosis in yeast: Evidence for the involvement of a small GTP-binding protein (Ypt7p). Cell 1992, 71, 1131–1142. [Google Scholar] [CrossRef]
- Chanda, A.; Roze, L.V.; Kang, S.; Artymovich, K.A.; Hicks, G.R.; Raikhel, N.V.; Calvo, A.M.; Linz, J.E. A key role for vesicles in fungal secondary metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 19533–19538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.; Zheng, W.; Wu, C.; Yang, J.; Xi, Y.; Xie, Q.; Zhao, X.; Deng, X.; Lu, G.; Li, G. Rab GTP ases are essential for membrane trafficking-dependent growth and pathogenicity in Fusarium graminearum. Environ. Microbiol. 2015, 17, 4580–4599. [Google Scholar] [CrossRef]
- Liu, X.-H.; Chen, S.-M.; Gao, H.-M.; Ning, G.-A.; Shi, H.-B.; Wang, Y.; Dong, B.; Qi, Y.-Y.; Zhang, D.-M.; Lu, G.-D.; et al. The small GTPase MoYpt7 is required for membrane fusion in autophagy and pathogenicity of Magnaporthe oryzae. Environ. Microbiol. 2015, 17, 4495–4510. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, M.; Cabrera, M.; Perz, A.; Bröcker, C.; Ostrowicz, C.; Engelbrecht-Vandré, S.; Ungermann, C. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 2010, 20, 1654–1659. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.-W.; Stromhaug, P.E.; Kauffman, E.J.; Weisman, L.S.; Klionsky, D.J. Yeast homotypic vacuole fusion requires the Ccz1-Mon1 complex during the tethering/docking stage. J. Cell Biol. 2003, 163, 973–985. [Google Scholar] [CrossRef]
- Haas, A.; Scheglmann, D.; Lazar, T.; Gallwitz, D.; Wickner, W. The GTPase Ypt7p of Saccharomyces cerevisiae is required on both partner vacuoles for the homotypic fusion step of vacuole inheritance. EMBO J. 1995, 14, 5258–5270. [Google Scholar] [CrossRef]
- Gao, H.-M.; Liu, X.-G.; Shi, H.-B.; Lu, J.-P.; Yang, J.; Lin, F.-C.; Liu, X.-H. MoMon1 is required for vacuolar assembly, conidiogenesis and pathogenicity in the rice blast fungus Magnaporthe oryzae. Res. Microbiol. 2013, 164, 300–309. [Google Scholar] [CrossRef]
- Desjardins, A.E.; Bai, G.-H.; Plattner, R.D.; Proctor, R.H. Analysis of aberrant virulence of Gibberella zeae following transformation-mediated complementation of a trichothecene-deficient (Tri5) mutant. Microbiology 2000, 146 Pt 8, 2059–2068. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Li, B.; Liu, L.; Chen, H.; Zhang, H.; Zheng, X.; Zhang, Z. FgMon1, a guanine nucleotide exchange factor of FgRab7, is important for vacuole fusion, autophagy and plant infection in Fusarium graminearum. Sci. Rep. 2015, 5, 18101. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Yang, J.; An, Y.; Pan, Y.; Liu, G. Over-expression of pcvA involved in vesicle-vacuolar fusion affects the conidiation and penicillin production in Penicillium chrysogenum. Biotechnol. Lett. 2012, 34, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Ohsumi, K.; Arioka, M.; Nakajima, H.; Kitamoto, K. Cloning and characterization of a gene (avaA) from Aspergillus nidulans encoding a small GTPase involved in vacuolar biogenesis. Gene 2002, 291, 77–84. [Google Scholar] [CrossRef]
- Wu, M.-D.; Cheng, M.-J.; Yech, Y.-J.; Chen, Y.-L.; Chen, K.-P.; Yang, P.-H.; Chen, I.-S.; Yuan, G.-F. Monascusazaphilones A-C, three new azaphilone analogues isolated from the fungus Monascus purpureus BCRC 38108. Nat. Prod. Res. 2013, 27, 1145–1152. [Google Scholar] [CrossRef]
- Liu, J.; Lei, M.; Zhou, Y.; Chen, F. A Comprehensive Analysis of the Small GTPases Ypt7 Involved in the Regulation of Fungal Development and Secondary Metabolism in Monascus ruber M7. Front. Microbiol. 2019, 10, 452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, K.; Hicks, J.K.; Huang, T.-P.; Keller, N.P. Pka, Ras and RGS protein interactions regulate activity of AflR, a Zn (II) 2Cys6 transcription factor in Aspergillus nidulans. Genetics 2003, 165, 1095–1104. [Google Scholar] [CrossRef] [PubMed]
- Kalleda, N.; Naorem, A.; Manchikatla, R.V. Targeting fungal genes by diced siRNAs: A rapid tool to decipher gene function in Aspergillus nidulans. PLoS ONE 2013, 8, e75443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labbaoui, H.; Bogliolo, S.; Ghugtyal, V.; Solis, N.V.; Filler, S.G.; Arkowitz, R.A.; Bassilana, M. Role of Arf GTPases in fungal morphogenesis and virulence. PLoS Pathog. 2017, 13, e1006205. [Google Scholar] [CrossRef] [Green Version]
- Kawada, D.; Kobayashi, H.; Tomita, T.; Nakata, E.; Nagano, M.; Siekhaus, D.E.; Toshima, J.Y.; Toshima, J. The yeast Arf-GAP Glo3p is required for the endocytic recycling of cell surface proteins. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2015, 1853, 144–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nordbring-Hertz, B. Morphogenesis in the nematode-trapping fungus Arthrobotrys oligospora—An extensive plasticity of infection structures. Mycologist 2004, 18, 125–133. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Yang, X.; Xie, M.; Zhang, G.; Yang, L.; Bai, N.; Zhao, Y.; Li, D.; Zhang, K.-Q.; Yang, J. The Arf-GAP AoGlo3 regulates conidiation, endocytosis, and pathogenicity in the nematode-trapping fungus Arthrobotrys oligospora. Fungal Genet. Biol. 2020, 138, 103352. [Google Scholar] [CrossRef]
- Yang, J.; Liang, L.; Li, J.; Zhang, K.-Q. Nematicidal enzymes from microorganisms and their applications. Appl. Microbiol. Biotechnol. 2013, 97, 7081–7095. [Google Scholar] [CrossRef] [PubMed]
- An, B.; Li, B.; Qin, G.; Tian, S. Function of small GTPase Rho3 in regulating growth, conidiation and virulence of Botrytis cinerea. Fungal Genet. Biol. 2015, 75, 46–55. [Google Scholar] [CrossRef]
- Heupel, S.; Roser, B.; Kuhn, H.; Lebrun, M.-H.; Villalba, F.; Requena, N. Erl1, a novel era-like GTPase from Magnaporthe oryzae, is required for full root virulence and is conserved in the mutualistic symbiont Glomus intraradices. Mol. Plant-Microbe Interactions 2010, 23, 67–81. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Li, G.; Zhang, S.; Jiang, C.; Qin, J.; Xu, J. Activation of the signalling mucin MoM sb2 and its functional relationship with C bp1 in Magnaporthe oryzae. Environ. Microbiol. 2015, 17, 2969–2981. [Google Scholar] [CrossRef] [PubMed]
- Mahlert, M.; Leveleki, L.; Hlubek, A.; Sandrock, B.; Bölker, M. Rac1 and Cdc42 regulate hyphal growth and cytokinesis in the dimorphic fungus Ustilago maydis. Mol. Microbiol. 2006, 59, 567–578. [Google Scholar] [CrossRef]
- Kayano, Y.; Tanaka, A.; Takemoto, D. Two closely related Rho GTPases, Cdc42 and RacA, of the en-dophytic fungus Epichloë festucae have contrasting roles for ROS production and symbiotic infection synchronized with the host plant. PLoS Pathog. 2018, 14, e1006840. [Google Scholar] [CrossRef] [Green Version]
- Nesher, I.; Minz, A.; Kokkelink, L.; Tudzynski, P.; Sharon, A. Regulation of pathogenic spore germination by CgRac1 in the fungal plant pathogen Colletotrichum gloeosporioides. Eukaryot. Cell 2011, 10, 1122–1130. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Wang, Y.; Ma, B.; Hou, J.; Jin, Y.; Zhang, Y.; Ke, X.; Tai, L.; Zuo, Y.; Dey, K. Clg2p interacts with Clf and ClUrase to regulate appressorium formation, pathogenicity and conidial morphology in Curvularia lunata. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, H.; Miao, P.; Lin, X.; Li, L.; Wu, C.; Chen, X.; Abubakar, Y.S.; Norvienyeku, J.; Li, G.; Zhou, J. Small GTPase Rab7-mediated FgAtg9 trafficking is essential for autophagy-dependent development and pathogenicity in Fusarium graminearum. PLoS Genet. 2018, 14, e1007546. [Google Scholar] [CrossRef] [PubMed]
- Aboelfotoh Hendy, A.; Xing, J.; Chen, X.; Chen, X. The farnesyltransferase β-subunit RAM1 regulates localization of RAS proteins and appressorium-mediated infection in Magnaporthe oryzae. Mol. Plant Pathol. 2019, 20, 1264–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dagdas, Y.F.; Yoshino, K.; Dagdas, G.; Ryder, L.S.; Bielska, E.; Steinberg, G.; Talbot, N.J. Septin-mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae. Science 2012, 336, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Lippincott, J.; Shannon, K.B.; Shou, W.; Deshaies, R.J.; Li, R. The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J. Cell Sci. 2001, 114, 1379–1386. [Google Scholar] [CrossRef]
- Sundaram, S.; Kim, S.J.; Suzuki, H.; Mcquattie, C.J.; Hiremah, S.T.; Podila, G.K. Isolation and characterization of a symbiosis-regulated ras from the ectomycorrhizal fungus Laccaria bicolor. Mol. Plant. Microbe. Interact. 2001, 14, 618–628. [Google Scholar] [CrossRef] [Green Version]
- Chiarugi, P.; Pani, G.; Giannoni, E.; Taddei, L.; Colavitti, R.; Raugei, G.; Symons, M.; Borrello, S.; Galeotti, T.; Ramponi, G. Reactive oxygen species as essential mediators of cell adhesion: The oxidative inhibition of a FAK tyrosine phosphatase is required for cell adhesion. J. Cell Biol. 2003, 161, 933–944. [Google Scholar] [CrossRef]
- Perotto, S.; Baluška, F. Signaling and Communication in Plant Symbiosis; Springer: Berlin/Heidelberg, Germany, 2012; ISBN 3642209653. [Google Scholar]
- Scott, B.; Green, K.; Berry, D. The fine balance between mutualism and antagonism in the Epichloë festucae—Grass symbiotic interaction. Curr. Opin. Plant Biol. 2018, 44, 32–38. [Google Scholar] [CrossRef]
- Segal, L.M.; Wilson, R.A. Reactive oxygen species metabolism and plant-fungal interactions. Fungal Genet. Biol. 2018, 110, 1–9. [Google Scholar] [CrossRef] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dautt-Castro, M.; Rosendo-Vargas, M.; Casas-Flores, S. The Small GTPases in Fungal Signaling Conservation and Function. Cells 2021, 10, 1039. https://doi.org/10.3390/cells10051039
Dautt-Castro M, Rosendo-Vargas M, Casas-Flores S. The Small GTPases in Fungal Signaling Conservation and Function. Cells. 2021; 10(5):1039. https://doi.org/10.3390/cells10051039
Chicago/Turabian StyleDautt-Castro, Mitzuko, Montserrat Rosendo-Vargas, and Sergio Casas-Flores. 2021. "The Small GTPases in Fungal Signaling Conservation and Function" Cells 10, no. 5: 1039. https://doi.org/10.3390/cells10051039