1. Introduction
Cells growing aerobically must deal with reactive oxygen species (ROS) as byproducts of respiration. These include (proceeding from most oxidized to most reduced) superoxide radicals, hydrogen peroxide, and hydroxyl radicals. These exhibit half-lives of ~5 s, days to weeks, and ~10
–9 s, respectively. The highly reactive and very short-lived hydroxyl radicals damage anything in their immediate vicinity. Enzymes exist in most cells to eliminate superoxide radicals (superoxide dismutase) and peroxides (glutathione peroxidase and catalases), but not the short-lived hydroxyl radicals. Conversion of the radical species to longer lived and diffusible H
2O
2 can lead to oxidation of sulfydryls and other groups, and also participate in the Fenton reaction with FeII. Fe-S clusters, with their important redox, catalytic, and regulatory functions, may be significant targets, as well as mononuclear iron proteins [
1,
2]. The Fenton reaction would not simply damage the Fe-S clusters but would also generate more hydroxyl radicals. These and related processes slowly damage cellular proteins and peptides and may contribute to aging in higher eukaryotes [
3,
4,
5,
6,
7,
8,
9,
10]. The connection between protein oxidation and aging has been a theme in the recent research from the laboratory of Miroslav Radman [
11,
12,
13,
14,
15], to which this issue and article are dedicated. The role of protein oxidation in aging remains an important focus of research [
3,
4,
5,
6,
7,
8,
9,
10].
Many questions remain to be answered about proteome damage via these oxidative processes. How fast does it occur? What proteins and parts of proteins are most susceptible to damage? How much of that damage can be repaired? There are many more. The effects of ionizing radiation (IR) on cellular proteomes can provide some important clues. IR such as gamma radiation (X-rays, 60Co decay, etc.) causes damage primarily via radiolysis of water and dissolved O2, generating ROS. High doses of IR can be administered in short amounts of time allowing investigators to rapidly increase intracellular levels of ROS and examine the effects of such an extreme stress.
In wild type
E. coli, an IR dose of 1000 Gy kills most of the cells. The proteome suffers relatively little damage [
16] but the genome is fragmented by IR-induced double-strand breaks (DSBs) [
16]. Cell death is most likely due to the DNA damage caused by ROS, including hydroxyl radicals from various sources including Fenton chemistry. However, damage to other cellular components, such as Fe-S complexes (also via Fenton chemistry) may play a role [
1,
2]. Additional oxidation events may cause enzyme inactivation and cell wall damage.
Using experimental evolution, we have been pursuing a long-term project to generate populations of
Escherichia coli that are as radiation resistant as the bacterium
Deinococcus radiodurans [
17,
18,
19,
20]. Progress reports have been published [
17,
18]. IR resistance has continued to increase. In addition to the evolution experiment, we have begun to catalogue proteome damage imposed by exposure to IR in both wildtype
E. coli and these evolved isolates. We previously focused on wild type
E. coli and
D. radiodurans, examining proteome damage inflicted by 1000 Gy. Whereas this dose is sufficient to kill more than 99.9% of
E. coli cells, it does not result in measurable lethality in
D. radiodurans [
16]. Limited proteome damage was noted in
E. coli (only 22 of 13,262 peptides detected were both oxidized and exhibited a fold increase in abundance >2) with little to no damage seen in
D. radiodurans. One major oxidation event, trioxidation of an active site Cys residue in glyceraldehyde 3-phosphate dehydrogenase (GAPDH), dominated the oxidative spectrum but was not present in
Deinococcus [
16]. In the present report, we examine proteome damage inflicted by much higher doses of IR. We compare
Escherichia coli and
Deinococcus radiodurans, as well as two isolates from highly evolved populations generated in the evolution trials [
17,
18]. Our goal was to continue cataloguing the types of protein oxidation events that occur as well as to begin to determine the extent to which IR-resistance is contingent on a capacity for amelioration of protein oxidation comparable to
Deinococcus. Using a dose of 3000 Gy, an increased level of damage is evident, allowing a more systematic examination of oxidative damage to bacterial proteomes.
3. Results
Previous work has often examined biological responses to IR exposure. Here, we wish to quantify the immediate, abiotic chemical effects of IR on the in vivo proteome, prior to any biological response. To ensure our observations of IR-induced changes to the
Escherichia coli proteome are indeed abiotic, cultures were cooled to 4 °C prior to irradiation. Unlike the previous study,
E. coli cells were grown in LB rich media throughout irradiation, due to diminished growth rate of the evolved isolates in media other than LB. However, all cells were extensively washed in 1X PBS to halt metabolism and remove nutrients in growth media that may act as a radioprotectant. In addition, sample tubes were submerged in cold (4–10 °C) water throughout irradiation. Samples were frozen in liquid nitrogen immediately after irradiation and remained frozen and protected from light and O
2 until processing. In this study, IR was delivered at 70 Gy/min by a high energy electron beam linear accelerator (Linac) commonly used in cancer radiotherapy. Under the conditions used in our experiments, a dose of 3000 Gy was administered quickly (~40 min). Based on previous measurements, this dose is sufficient to cause approximately 60 DNA DSBs per cell, and is also expected to induce significant oxidative damage to proteins [
27].
In order to provide several comparative proteomes from IR-resistant species, we carried out identical experiments as laid out above with (a) two isolates from a population that had been subjected to 150 rounds of selection for IR resistance as part of a long-term evolution trial to generate high levels of IR resistance [
17,
18] and (b) the highly radioresistant bacterium
Deinococcus radiodurans. The isolates from the experimental evolution trial are both from one population (IR9) and are denoted IR9-150-1 and IR9-150-2. They are from two different sub-populations within IR9 that are involved in persistent clonal interference [
18]. The ability of
Deinococcus to protect its proteome from IR-generated ROS is a well-studied phenomenon [
27,
28,
29,
30]. At a dose of 3000 Gy, this bacterium exhibits no lethality when subjected to a slower dose rate of IR up to 5000 Gy [
30,
31,
32,
33,
34,
35]. However, a dose of 3000 Gy administered from the Linac kills 50–90% of the cells in a culture of
D. radiodurans [
17,
18]. Survival of the
E. coli isolates subjected to experimental evolution is approaching that of
D. radiodurans [
17,
18]. The dose of 3000 Gy was chosen to provide a broader spectrum of proteome damage than that catalogued in an earlier study [
16].
Samples were thawed and processed immediately. To survey and quantify as much IR-induced oxidation as possible, peptides from ten replicates (five treated and five mock-treated) were analyzed using mass spectrometry (MS). The proteome databases were searched systematically for previously identified IR-induced amino acid modifications, including carbonylation (+13.98 Da), hydroxylation (+15.99 Da), dioxidation (+31.99 Da), and trioxidation (+47.99 Da) [
26,
36]. We note here that although we classify +31.99 Da as dioxidation, such a mass shift could also be peroxidation. Though these modifications represent different chemistries, we cannot distinguish between the two in these data. We also note that we are using general terms for modification here, though more specific terminology will be used in some instances (i.e., +47.99 Da on Cys results in a sulfonic acid chemical group, +13.98 Da on Pro can represent conversion to pyroglutamic acid, etc.). Given their scarcity in previous work, we did not search for additional previously reported but less-common modifications [
26,
36,
37].
The coverage in our datasets is summarized in
Table 1,
Table 2,
Table 3 and
Table 4. The total number of peptides detected ranged from 14,738 to 21,516 in the samples obtained from the three
E. coli strains and 14,440 from
D. radiodurans, respectively. Unique proteins detected in each strain were: MG1655, 2020; IR9-150-1, 1868; IR9-150-2, 1703;
D. radiodurans, 954. This corresponded to nearly half of the predicted proteome for each strain, or most of the proteins typically expressed during exponential phase growth [
38]. Of the peptides detected, a relatively small fraction contained oxidative modifications (
Table 1,
Table 2,
Table 3 and
Table 4). The highlighting added to draw attention to the relative occurrence of major oxidation classes.
We tabulated the peptides that exhibited either changes or no change in abundance in the irradiated samples relative to the unirradiated samples, focusing first on the vast majority that exhibited no statistically significant change in abundance after IR treatment at 3000 Gy (
Table 1,
Table 2,
Table 3 and
Table 4). Most of these peptides were unmodified. The overall patterns of detected peptides are shown in
Figure 1.
A subset of the peptides that exhibited no changes in abundance were modified. There are two potential sources contributing to a background of oxidatively modified peptides that do not change in abundance due to IR exposure, endogenous intracellular ROS species or source oxidation during electrospray ionization. Since we froze the samples in this study immediately after irradiation and protected them from the environment, the involvement of endogenous ROS should be reduced relative to a previous study [
16]. The observed background of oxidized peptides is similar in both
E. coli and
D. radiodurans.
D. radiodurans has a documented enhanced capacity to ameliorate ROS [
29,
39]. If we assume that
D. radiodurans is better able to suppress intracellular ROS than is
E. coli, yet the background of oxidatively modified peptides is similar or even higher than in
E. coli, the likely origin of our oxidized peptide background is in-source oxidation during electrospray ionization of peptides. Determining the precise source of these IR-independent modifications is a continuing effort but is outside the focus of this study. The background modification is in our whole cell samples and is consistent with previous observations of endogenous ROS produced during growth in nutrient-rich medium [
40,
41] and oxidation of proteins during the electrospray ionization step of MS [
42]. For the subset of oxidized peptides that showed no change in abundance, single hydroxylation (+15.99 Da), especially on methionine, was the most prevalent modification (~7–12%) (
Table 5).
The other remaining peptides exhibited a statistically significant increase or decrease in abundance (fold change greater than 2, adjusted
p-value < 0.05) (
Table 1,
Table 2,
Table 3 and
Table 4). Again, most of these peptides were unmodified. In principle, changes in abundance of unmodified peptides can occur in at least three ways. There could be an increase in synthesis, increase or decrease in proteolysis, both increase in synthesis and a change in proteolysis. Changes to proteolysis would reflect changes in access to trypsin cleavage sites. The extraction procedure, which involves the use of detergents, should eliminate most protein–protein interactions, so changes in association should not unduly affect our access to peptides. However, in principle, any interactions not eliminated by our protocol might be affected instead by the irradiation, leading to increased access to trypsin cleavage sites and an observed increase in certain peptides. Our attempts to suppress cell metabolism during irradiation may not be perfect. In spite of the absence of growth and nutrients, as well as the cold temperature to halt metabolism, certain proteins may increase in abundance during the 40 min irradiation process. Alternatively, modification or damage to a peptide in one part of the protein may lead to a conformation change that leads to elevated trypsin cleavage of otherwise occluded sites in the protein to produce a corresponding increase in the amount of a corresponding undamaged peptide. Overall, about 75–95% of the peptides with increases or decreases in abundance were unmodified in each
E. coli strain tested (
Table 1,
Table 2 and
Table 3). Far fewer peptides changed in abundance in
D. radiodurans, suggestive of the powerful effect of enhanced ROS amelioration on suppressing oxidative protein damage. However, of those few peptides, a larger fraction were oxidized (nearly 50%) (
Figure 1,
Table 4).
The oxidation events that occurred on peptides that were modified and also increased or decreased in abundance are listed in
Table 6 and
Table 7. The peptides of most interest are those that both exhibit an increase in abundance and are oxidatively modified. We associate significant increases in the abundance (fold change greater than two) of oxidatively modified peptides with the primary effect of irradiation. The total number of modified peptides in this class increased somewhat from our previous study, although perhaps not as much as might be expected due to the 3X increased IR dose used in this study. The overall fraction of the peptides increasing in abundance that were oxidized varied among the four samples. This number ranged from 40% in wild type
E. coli to 18% and 17% in the evolved isolates of
E. coli to 78% for
D. radiodurans. Note that only 17 peptides fall into this category in
Deinococcus. Again, hydroxylation, especially hydroxylation of methionine, was the most common oxidation event observed (
Table 6).
We further investigated the level of oxidation on peptides that appeared only in the irradiated sample. Although such peptides could not be assigned a quantitative fold increase, an oxidized peptide that appeared only in irradiated cells likely represents the most significant form of IR-induced protein oxidation. Nearly 30% of peptides in this category were oxidized in all organisms tested (
Figure 1). Again, the most common oxidation event, by a considerable margin, was hydroxylation. Carbonylation was second, followed by rarer deoxidation and trioxidation events. This was true in all samples.
Overall, there were 249 peptides from WT E. coli that were both oxidized and present at increased abundance (fold increase greater than two). This number declined to 106 and 65 for IR9-150-1 and IR9-150-2, respectively, suggesting the possible presence of increased ROS amelioration that is adding proteomic protection in these cells. In D. radiodurans, 50 peptides with oxidative modifications and increased abundance were detected. Thus, the result with IR9-150-2 suggests that if amelioration of proteome oxidation is present, it may be comparable to that seen in D. radiodurans. The total number of proteins represented among these oxidized peptides was 157 for WT E. coli, 81 for IR9-150-1, 44 IR9-150-2, and only 38 for D. radiodurans.
While such rampant oxidation appears scattershot, we sought to determine what proteins (if any) were more susceptible to IR induced damage than others. We narrowed our analysis to any proteins with at least two oxidized peptides with increased abundance in the irradiated samples (quantified fold change or present in the irradiated sample only). Of the 157 total proteins from the MG1655 dataset that had modified peptides that increased in abundance, only 35 had multiple peptides with modifications (
Figure 2). In some cases, as in the four proteins at the far left, all of the peptides detected in both unirradiated and irradiated samples exhibited a similar increase in abundance in the irradiated samples. This suggests that the protein itself increased in abundance during the irradiation. Much more commonly, most of the peptides derived from a particular protein exhibit no change in abundance. Most of these peptides are unmodified, whereas a few peptides from the same protein exhibit increases in abundance. Most of the latter peptides were modified. These are generally proteins that did not change in abundance during the irradiation process, although some of the peptides from those proteins were modified and those modified versions of the peptides exhibited an increase in the irradiated samples.
Figure 2 focuses on proteins where multiple modifications were detected, but most of the modified proteins (517 total) had no peptides that increased in abundance (394 such proteins), or only one modified peptide that increased in abundance (123 proteins).
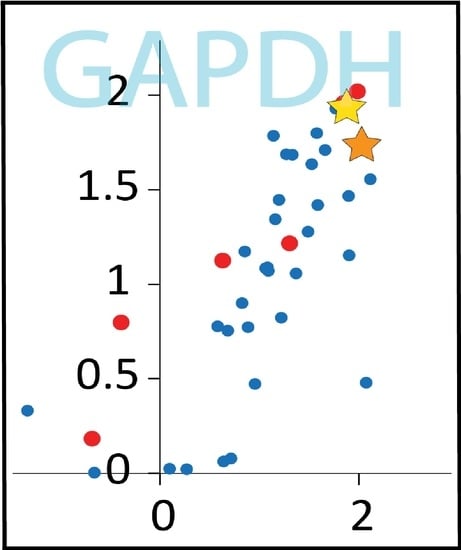
Of the proteins displayed in
Figure 2, four have multiple oxidized peptides that appeared only in the irradiated samples: GAPDH, elongation factor (Ef)-Tu (TufA), Ef-G (FusA), and the ribosomal protein RplP. These four proteins may represent the most prominent targets of IR-induced protein oxidation. Further analysis revealed that GAPDH, Ef-Tu, and Ef-G are also clear targets of IR-induced oxidation in IR9-150-1 and IR9-150-2 (
Figure 3).
We have previously noted the IR-sensitivity of GAPDH, Ef-Tu, and Ef-G [
16], and the presence of increased oxidation on these proteins in even IR-resistant
E. coli isolates highlights their susceptibility.
In particular, only GAPDH and Ef-Tu have modified peptides (three in GAPDH and one in Ef-Tu) that increased in abundance greater than two fold in all three
E. coli strains (
Figure 3, denoted by stars). For GAPDH, one of these peptides was derived from the active site and containing the active site Cys residue, which is subject to trioxidation [
16]. In the current study, this GAPDH oxidation event could only be localized to the active site Cys in MG1655, but the mass shifts for the oxidized active site peptide in the evolved isolates are consistent with oxidation of Cys to sulfonic acid (+48 Da). Incomplete oxidation (dioxidation, +32 Da) is also apparent (
Supplemental Tables S1–S3). The sequence of this active site peptide is highly conserved in bacteria and much of it is conserved in other organisms. This same peptide does not suffer detectable oxidation in the GAPDH from
D. radiodurans, even at 3000 Gy. In this gene,
D. radiodurans (and the rest of the Deinococcus-Thermus phylum) has a rare sequence alteration that eliminates a second Cys residue that is part of the normal active site consensus for this enzyme [
43], a change that may render it much less sensitive to oxidation. It has been previously suggested that the ROS-sensitivity of GAPDH may act as a metabolic switch necessary to shift carbon flux to the pentose phosphate pathway to generate NADPH used in reducing glutathione and in biosynthesis [
43,
44,
45,
46]. Two more modified peptides from GAPDH were increased in abundance in all three
E. coli isolates. These peptides map to elsewhere in the protein, and oxidation is likely localized to
l-met residues (M129 and M229) in all three isolates (localization to
l-met is confirmed in IR9-150-1 and IR9-150-2, but inconclusive in MG1655).
Less evident in earlier work has been the effects of IR on Ef-Tu. A residue of Ef-Tu, F47, may be particularly ROS-sensitive as revealed in this new dataset. This residue is quite close to the GDP binding site. An IR-induced and very prominent hydroxylation event was localized to F47 in a peptide from IR9-150-2 (22-fold increase). Mass shifts corresponding to hydroxylation were detected in the same peptide at increased levels in MG1655 (10-fold increase) and IR9-150-1 (4-fold increase) (
Figure 3; see also
Figure S1), although the modified residue could not be unambiguously identified in the latter two strains. To our knowledge, oxidation sensitivity of this residue has not been previously reported and the biological consequence of such modification is unknown.
In
Deinococcus radiodurans, only one protein had multiple peptides that both were oxidized and increased in abundance. This is an SLH protein, part of the S layer on the cell surface, which has three oxidized peptides that increase in abundance. These oxidized peptides were detected only in the irradiated sample. Another protein that is part of the S layer, SlpA, provided one peptide that was the only measurable target of IR-mediated oxidation in our previous trial carried out at 1000 Gy [
16]. SlpA also has one oxidized peptide in the current dataset, although the increase in abundance of that peptide is not as great as the three peptides from the SLH protein.
Deinococcus may not be able to protect cell surface proteins from IR-mediated oxidation as well as proteins in the cell interior. The prominent oxidation events in
Deinococcus are summarized in
Table 8. The entire dataset is provided in
Supplemental Table S4. The peptide from SlpA is not included as the increase in abundance is modest.
4. Discussion
The work presented in this study leads to multiple conclusions. First, of the oxidation events detected, hydroxylation is the most prominent. Many previous studies have focused efforts on detecting carbonylation and this has been reported to be the main oxidation event affecting proteins [
14,
47,
48]. This and our previous work [
16] indicates that much more attention should be focused on hydroxylation events. Second, the overall level of proteomic oxidation is not extensive, even at 3000 Gy. There were only 480 modified peptides from all four samples that exhibited an increase in abundance of > 2-fold. Even in WT
E. coli, only 157 proteins, out of 2020 detected, exhibited any detectable modification that was clearly attributable to IR. Of these, only 35 had multiple peptides that were so modified. Finally, glyceraldehyde phosphate dehydrogenase again figures prominently in the modification dataset. The new dataset also provided a closer look at Ef-Tu as a major oxidation target.
The proteins that have only one or a few modified peptides that increase in abundance vary from one
E. coli strain to another but are always few in number. The observed increases are also often small. This may indicate that even with an IR dose of 3000 Gy, we are far from saturation relative to the proteomic oxidative events that might occur. In wild type
E. coli, the proteome is remarkably resistant to oxidation as long as cells remain intact [
16]. The higher IR doses in the present study highlight the presence of some proteins where there are oxidatively modified peptides that increase substantially in abundance while other peptides in the same protein are not modified and do not change in abundance. This suggests that the limited protein oxidation that does occur may be greatly affected by factors such as protein structure and exposure to water, as has also been seen in studies of IR effects on individual proteins such as lysozyme [
11].
The presence of a few highly sensitive targets of oxidation appearing in all
E. coli samples, such as the active site peptide of glyceraldehyde 3-phosphate dehydrogenase and one peptide of Ef-Tu, suggest that proteome oxidation is not at all random. There may be an evolutionary component to protein oxidation potential. Targeted oxidation of these particular peptides in GAPDH and Ef-Tu may improve survival under conditions of oxidative stress. Inactivation of GAPDH directly slows glycolysis, funneling hexose metabolism into the pentose phosphate pathway. The resulting production of NADPH can play a role in ROS amelioration, providing reduced glutathione for glutathione peroxidase [
49,
50]. The active site of GAPDH is widely conserved, suggesting that an ROS amelioration mechanism of this kind is present in cells from bacteria to humans. Inactivation or partial inactivation of Ef-Tu may also be an adaptive response that improves survival to oxidative damage. Protein biosynthesis typically consumes 80–90% of the chemical energy resources of a cell (ATP; [
51]). In principle, a transient slowdown of this process could free resources to deal with DNA damage as well as damage to Fe-sulfur centers and other cellular components. In principle, lower translation rates might also preserve pools of intracellular GTP for repairing oxidative damage to DNA.
The study of proteome oxidation is still in its infancy. As with many of the fields represented in this Special Issue of
Cells, Miroslav Radman has made early and prominent contributions, pointing out the potential importance of these processes to human health and aging [
11,
12,
13,
14,
15]. It is a privilege to dedicate this article to Miro on this auspicious occasion.