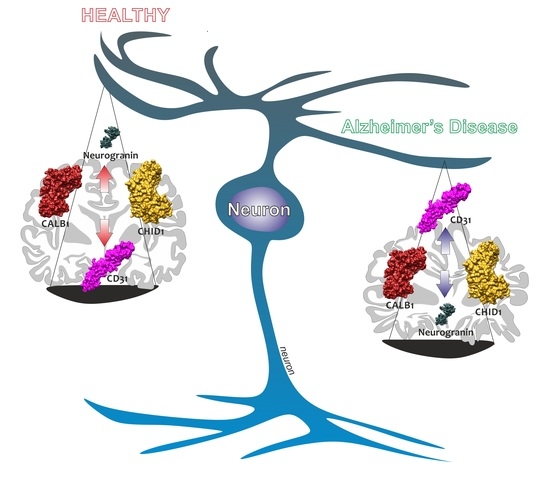
Brain CHID1 Expression Correlates with NRGN and CALB1 in Healthy Subjects and AD Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Selection
2.2. Clinical and Neuropathological Criteria
2.3. Data Processing and Experimental Design
2.4. Statistical Analysis
3. Results
3.1. Sex-Dependent Differences in CHID1 Brain Expression of NDHC Subjects and AD Patients
3.2. Neurodegeneration Markers Correlated with CHID1 Expression Levels in the CNS
3.3. CHID1 Levels Are Differentially Expressed in Eight Brain Regions
3.4. ROC Analysis Confirmed the Diagnostic Ability of CHID1 to Discriminate AD Patients from NDHC Subjects
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gitler, A.D.; Dhillon, P.; Shorter, J. Neurodegenerative disease: Models, mechanisms, and a new hope. Dis. Models Mech. 2017, 10, 499–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef]
- Blennow, K.; de Leon, M.J.; Zetterberg, H. Alzheimer’s disease. Lancet 2006, 368, 387–403. [Google Scholar] [CrossRef]
- Kaya, I.; Brinet, D.; Michno, W.; Baskurt, M.; Zetterberg, H.; Blenow, K.; Hanrieder, J. Novel Trimodal MALDI Imaging Mass Spectrometry (IMS3) at 10 mum Reveals Spatial Lipid and Peptide Correlates Implicated in Abeta Plaque Pathology in Alzheimer’s Disease. ACS Chem. Neurosci. 2017, 8, 2778–2790. [Google Scholar] [CrossRef]
- Frankel, R.; Tornquist, M.; Meisl, G.; Hansson, O.; Andreasson, U.; Zetterberg, H.; Blennow, K.; Frohm, B.; Cedervall, T.; Knowles, T.P.J.; et al. Autocatalytic amplification of Alzheimer-associated Abeta42 peptide aggregation in human cerebrospinal fluid. Commun. Biol. 2019, 2, 365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanfilippo, C.; Longo, A.; Lazzara, F.; Cambria, D.; Distefano, G.; Palumbo, M.; Cantarella, A.; Malaguarnera, L.; Di Rosa, M. CHI3L1 and CHI3L2 overexpression in motor cortex and spinal cord of sALS patients. Mol. Cell. Neurosci. 2017, 85, 162–169. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, X.; Yuan, X.; Hao, L. Role of allograft inflammatory factor-1 in the regulation of inflammation and oxidative stress in primary peritoneal mesothelial cells. Cell Biol. Int. 2019, 43, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Kalinowska, A.; Losy, J. PECAM-1, a key player in neuroinflammation. Eur. J. Neurol. 2006, 13, 1284–1290. [Google Scholar] [CrossRef]
- Fairless, R.; Williams, S.K.; Diem, R. Calcium-Binding Proteins as Determinants of Central Nervous System Neuronal Vulnerability to Disease. Int. J. Mol. Sci 2019, 20, 2146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Rosa, M.; De Gregorio, C.; Malaguarnera, G.; Tuttobene, M.; Biazzo, F.; Malaguarnera, L. Evaluation of AMCase and CHIT-1 expression in monocyte macrophages lineage. Mol. Cell. Biochem. 2013, 374, 73–80. [Google Scholar] [CrossRef]
- Di Rosa, M.; Tibullo, D.; Cambria, D.; Distefano, G.; Saccone, S.; Di Raimondo, F.; Malaguarnera, L. Chitotriosidase Expression during Monocyte-Derived Dendritic Cells Differentiation and Maturation. Inflammation 2015, 38, 2082–2091. [Google Scholar] [CrossRef]
- Sanfilippo, C.; Castrogiovanni, P.; Imbesi, R.; Kazakowa, M.; Musumeci, G.; Blennow, K.; Zetterberg, H.; Di Rosa, M. Sex difference in CHI3L1 expression levels in human brain aging and in Alzheimer’s disease. Brain Res. 2019, 1720, 146305. [Google Scholar] [CrossRef]
- Sanfilippo, C.; Malaguarnera, L.; Di Rosa, M. Chitinase expression in Alzheimer’s disease and non-demented brains regions. J. Neurol. Sci. 2016, 369, 242–249. [Google Scholar] [CrossRef]
- Di Rosa, M.; Sanfilippo, C.; Libra, M.; Musumeci, G.; Malaguarnera, L. Different pediatric brain tumors are associated with different gene expression profiling. Acta Histochem. 2015, 117, 477–485. [Google Scholar] [CrossRef]
- Di Rosa, M.; Malaguarnera, G.; De Gregorio, C.; Drago, F.; Malaguarnera, L. Evaluation of CHI3L-1 and CHIT-1 expression in differentiated and polarized macrophages. Inflammation 2013, 36, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Di Rosa, M.; Tibullo, D.; Saccone, S.; Distefano, G.; Basile, M.S.; Di Raimondo, F.; Malaguarnera, L. CHI3L1 nuclear localization in monocyte derived dendritic cells. Immunobiology 2016, 221, 347–356. [Google Scholar] [CrossRef]
- Di Rosa, M.; Tibullo, D.; Vecchio, M.; Nunnari, G.; Saccone, S.; Di Raimondo, F.; Malaguarnera, L. Determination of chitinases family during osteoclastogenesis. Bone 2014, 61, 55–63. [Google Scholar] [CrossRef]
- Qiu, Q.C.; Wang, L.; Jin, S.S.; Liu, G.F.; Liu, J.; Ma, L.; Mao, R.F.; Ma, Y.Y.; Zhao, N.; Chen, M.; et al. CHI3L1 promotes tumor progression by activating TGF-beta signaling pathway in hepatocellular carcinoma. Sci. Rep. 2018, 8, 15029. [Google Scholar] [CrossRef]
- Di Rosa, M.; Zambito, A.M.; Marsullo, A.R.; Li Volti, G.; Malaguarnera, L. Prolactin induces chitotriosidase expression in human macrophages through PTK, PI3-K., and MAPK pathways. J. Cell. Biochem. 2009, 107, 881–889. [Google Scholar] [CrossRef]
- Di Rosa, M.; Malaguarnera, L. Genetic variants in candidate genes influencing NAFLD progression. J. Mol. Med. 2012, 90, 105–118. [Google Scholar] [CrossRef]
- Di Rosa, M.; Malaguarnera, L. Chitinase 3 Like-1: An Emerging Molecule Involved in Diabetes and Diabetic Complications. Pathobiology 2016, 83, 228–242. [Google Scholar] [CrossRef]
- Di Rosa, M.; Szychlinska, M.A.; Tibullo, D.; Malaguarnera, L.; Musumeci, G. Expression of CHI3L1 and CHIT1 in osteoarthritic rat cartilage model. A morphological study. Eur. J. Histochem. EJH 2014, 58, 2423. [Google Scholar] [CrossRef] [Green Version]
- Kwak, E.J.; Hong, J.Y.; Kim, M.N.; Kim, S.Y.; Kim, S.H.; Park, C.O.; Kim, K.W.; Lee, C.G.; Elias, J.A.; Jee, H.M.; et al. Chitinase 3-like 1 drives allergic skin inflammation via Th2 immunity and M2 macrophage activation. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2019, 49, 1464–1474. [Google Scholar] [CrossRef]
- Kzhyshkowska, J.; Gratchev, A.; Goerdt, S. Stabilin-1, a homeostatic scavenger receptor with multiple functions. J. Cell. Mol. Med. 2006, 10, 635–649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanfilippo, C.; Nunnari, G.; Calcagno, A.; Malaguarnera, L.; Blennow, K.; Zetterberg, H.; Di Rosa, M. The chitinases expression is related to Simian Immunodeficiency Virus Encephalitis (SIVE) and in HIV encephalitis (HIVE). Virus Res. 2017, 227, 220–230. [Google Scholar] [CrossRef] [Green Version]
- Fagone, P.; Nunnari, G.; Lazzara, F.; Longo, A.; Cambria, D.; Distefano, G.; Palumbo, M.; Nicoletti, F.; Malaguarnera, L.; Di Rosa, M. Induction of OAS gene family in HIV monocyte infected patients with high and low viral load. Antivir. Res. 2016, 131, 66–73. [Google Scholar] [CrossRef]
- Thul, P.J.; Lindskog, C. The human protein atlas: A spatial map of the human proteome. Protein Sci. Publ. Protein Soc. 2018, 27, 233–244. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.F.; Ho, D.W.; Lau, C.K.; Lam, C.T.; Lum, C.T.; Poon, R.T.; Fan, S.T. Allograft inflammatory factor-1 (AIF-1) is crucial for the survival and pro-inflammatory activity of macrophages. Int. Immunol. 2005, 17, 1391–1397. [Google Scholar] [CrossRef] [PubMed]
- Del Galdo, F.; Maul, G.G.; Jimenez, S.A.; Artlett, C.M. Expression of allograft inflammatory factor 1 in tissues from patients with systemic sclerosis and in vitro differential expression of its isoforms in response to transforming growth factor beta. Arthritis Rheum. 2006, 54, 2616–2625. [Google Scholar] [CrossRef]
- Reus, G.Z.; Silva, R.H.; de Moura, A.B.; Presa, J.F.; Abelaira, H.M.; Abatti, M.; Vieira, A.; Pescador, B.; Michels, M.; Ignacio, Z.M.; et al. Early Maternal Deprivation Induces Microglial Activation, Alters Glial Fibrillary Acidic Protein Immunoreactivity and Indoleamine 2,3-Dioxygenase during the Development of Offspring Rats. Mol. Neurobiol. 2019, 56, 1096–1108. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, A.; Kawahito, Y. The immunologic function and role of allograft inflammatory factor-1. Nihon Rinsho Men’eki Gakkai Kaishi Jpn. J. Clin. Immunol. 2014, 37, 139–145. [Google Scholar] [CrossRef] [Green Version]
- Schwab, J.M.; Frei, E.; Klusman, I.; Schnell, L.; Schwab, M.E.; Schluesener, H.J. AIF-1 expression defines a proliferating and alert microglial/macrophage phenotype following spinal cord injury in rats. J. Neuroimmunol. 2001, 119, 214–222. [Google Scholar] [CrossRef]
- Giorgetti, E.; Panesar, M.; Zhang, Y.; Joller, S.; Ronco, M.; Obrecht, M.; Lambert, C.; Accart, N.; Beckmann, N.; Doelemeyer, A.; et al. Modulation of Microglia by Voluntary Exercise or CSF1R Inhibition Prevents Age-Related Loss of Functional Motor Units. Cell Rep. 2019, 29, 1539–1554.e7. [Google Scholar] [CrossRef]
- Privratsky, J.R.; Newman, P.J. PECAM-1: Regulator of endothelial junctional integrity. Cell Tissue Res. 2014, 355, 607–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wimmer, I.; Tietz, S.; Nishihara, H.; Deutsch, U.; Sallusto, F.; Gosselet, F.; Lyck, R.; Muller, W.A.; Lassmann, H.; Engelhardt, B. PECAM-1 Stabilizes Blood-Brain Barrier Integrity and Favors Paracellular T-Cell Diapedesis Across the Blood-Brain Barrier During Neuroinflammation. Front. Immunol. 2019, 10, 711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hochstrasser, T.; Weiss, E.; Marksteiner, J.; Humpel, C. Soluble cell adhesion molecules in monocytes of Alzheimer’s disease and mild cognitive impairment. Exp. Gerontol. 2010, 45, 70–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagawa, S.; Castro, V.; Toborek, M. Infection of human pericytes by HIV-1 disrupts the integrity of the blood-brain barrier. J. Cell. Mol. Med. 2012, 16, 2950–2957. [Google Scholar] [CrossRef]
- Limaos, E.A.; Borges, D.R.; Souza-Pinto, J.C.; Gordon, A.H.; Prado, J.L. Acute turpentine inflammation and kinin release in rat-paw thermic oedema. Br. J. Exp. Pathol. 1981, 62, 591–594. [Google Scholar]
- Kiyama, H.; Seto-Ohshima, A.; Emson, P.C. Calbindin D28K as a marker for the degeneration of the striatonigral pathway in Huntington’s disease. Brain Res. 1990, 525, 209–214. [Google Scholar] [CrossRef]
- Yuan, H.H.; Chen, R.J.; Zhu, Y.H.; Peng, C.L.; Zhu, X.R. The neuroprotective effect of overexpression of calbindin-D(28k) in an animal model of Parkinson’s disease. Mol. Neurobiol. 2013, 47, 117–122. [Google Scholar] [CrossRef]
- Kook, S.Y.; Jeong, H.; Kang, M.J.; Park, R.; Shin, H.J.; Han, S.H.; Son, S.M.; Song, H.; Baik, S.H.; Moon, M.; et al. Crucial role of calbindin-D28k in the pathogenesis of Alzheimer’s disease mouse model. Cell Death Differ. 2014, 21, 1575–1587. [Google Scholar] [CrossRef]
- Masliah, E.; Ge, N.; Achim, C.L.; Wiley, C.A. Differential vulnerability of calbindin-immunoreactive neurons in HIV encephalitis. J. Neuropathol. Exp. Neurol. 1995, 54, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Dumurgier, J.; Schraen, S.; Gabelle, A.; Vercruysse, O.; Bombois, S.; Laplanche, J.L.; Peoc’h, K.; Sablonniere, B.; Kastanenka, K.V.; Delaby, C.; et al. Cerebrospinal fluid amyloid-beta 42/40 ratio in clinical setting of memory centers: A multicentric study. Alzheimer’s Res. Ther. 2015, 7, 30. [Google Scholar] [CrossRef] [Green Version]
- Diez-Guerra, F.J. Neurogranin, a link between calcium/calmodulin and protein kinase C signaling in synaptic plasticity. IUBMB Life 2010, 62, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Singec, I.; Knoth, R.; Ditter, M.; Volk, B.; Frotscher, M. Neurogranin is expressed by principal cells but not interneurons in the rodent and monkey neocortex and hippocampus. J. Comp. Neurol. 2004, 479, 30–42. [Google Scholar] [CrossRef]
- Colom-Cadena, M.; Spires-Jones, T.; Zetterberg, H.; Blennow, K.; Caggiano, A.; DeKosky, S.T.; Fillit, H.; Harrison, J.E.; Schneider, L.S.; Scheltens, P.; et al. The clinical promise of biomarkers of synapse damage or loss in Alzheimer’s disease. Alzheimer’s Res. Ther. 2020, 12, 21. [Google Scholar] [CrossRef]
- Davidsson, P.; Blennow, K. Neurochemical dissection of synaptic pathology in Alzheimer’s disease. Int. Psychogeriatr. 1998, 10, 11–23. [Google Scholar] [CrossRef]
- Reddy, P.H.; Mani, G.; Park, B.S.; Jacques, J.; Murdoch, G.; Whetsell, W., Jr.; Kaye, J.; Manczak, M. Differential loss of synaptic proteins in Alzheimer’s disease: Implications for synaptic dysfunction. J. Alzheimer’s Dis. JAD 2005, 7, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Hellwig, K.; Kvartsberg, H.; Portelius, E.; Andreasson, U.; Oberstein, T.J.; Lewczuk, P.; Blennow, K.; Kornhuber, J.; Maler, J.M.; Zetterberg, H.; et al. Neurogranin and YKL-40: Independent markers of synaptic degeneration and neuroinflammation in Alzheimer’s disease. Alzheimer’s Res. Ther. 2015, 7, 74. [Google Scholar] [CrossRef] [Green Version]
- Sjostedt, E.; Zhong, W.; Fagerberg, L.; Karlsson, M.; Mitsios, N.; Adori, C.; Oksvold, P.; Edfors, F.; Limiszewska, A.; Hikmet, F.; et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 2020, 367. [Google Scholar] [CrossRef]
- Narayanan, M.; Huynh, J.L.; Wang, K.; Yang, X.; Yoo, S.; McElwee, J.; Zhang, B.; Zhang, C.; Lamb, J.R.; Xie, T.; et al. Common dysregulation network in the human prefrontal cortex underlies two neurodegenerative diseases. Mol. Syst. Biol. 2014, 10, 743. [Google Scholar] [CrossRef]
- Kumaran, R.; Cookson, M.R. Pathways to Parkinsonism Redux: Convergent pathobiological mechanisms in genetics of Parkinson’s disease. Hum. Mol. Genet. 2015, 24, R32–R44. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Meng, Q.; Xia, Y.; Ding, C.; Wang, L.; Dai, R.; Cheng, L.; Gunaratne, P.; Gibbs, R.A.; Min, S.; et al. The transcription factor POU3F2 regulates a gene coexpression network in brain tissue from patients with psychiatric disorders. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef] [Green Version]
- Gibbs, J.R.; van der Brug, M.P.; Hernandez, D.G.; Traynor, B.J.; Nalls, M.A.; Lai, S.L.; Arepalli, S.; Dillman, A.; Rafferty, I.P.; Troncoso, J.; et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010, 6, e1000952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Gaiteri, C.; Bodea, L.G.; Wang, Z.; McElwee, J.; Podtelezhnikov, A.A.; Zhang, C.; Xie, T.; Tran, L.; Dobrin, R.; et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 2013, 153, 707–720. [Google Scholar] [CrossRef] [Green Version]
- Hernandez, D.G.; Nalls, M.A.; Moore, M.; Chong, S.; Dillman, A.; Trabzuni, D.; Gibbs, J.R.; Ryten, M.; Arepalli, S.; Weale, M.E.; et al. Integration of GWAS SNPs and tissue specific expression profiling reveal discrete eQTLs for human traits in blood and brain. Neurobiol. Dis. 2012, 47, 20–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trabzuni, D.; Ramasamy, A.; Imran, S.; Walker, R.; Smith, C.; Weale, M.E.; Hardy, J.; Ryten, M.; North American Brain Expression, C. Widespread sex differences in gene expression and splicing in the adult human brain. Nat. Commun. 2013, 4, 2771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, H.; Hodges, A.K.; Curtis, C.; Lee, S.H.; Troakes, C.; Dobson, R.J.B.; Newhouse, S.J. Transcriptomic analysis of probable asymptomatic and symptomatic alzheimer brains. Brain Behav. Immun. 2019, 80, 644–656. [Google Scholar] [CrossRef]
- Kang, H.J.; Kawasawa, Y.I.; Cheng, F.; Zhu, Y.; Xu, X.; Li, M.; Sousa, A.M.; Pletikos, M.; Meyer, K.A.; Sedmak, G.; et al. Spatio-temporal transcriptome of the human brain. Nature 2011, 478, 483–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- French, L.; Ma, T.; Oh, H.; Tseng, G.C.; Sibille, E. Age-Related Gene Expression in the Frontal Cortex Suggests Synaptic Function Changes in Specific Inhibitory Neuron Subtypes. Front. Aging Neurosci. 2017, 9, 162. [Google Scholar] [CrossRef] [Green Version]
- Ryan, M.M.; Lockstone, H.E.; Huffaker, S.J.; Wayland, M.T.; Webster, M.J.; Bahn, S. Gene expression analysis of bipolar disorder reveals downregulation of the ubiquitin cycle and alterations in synaptic genes. Mol. Psychiatry 2006, 11, 965–978. [Google Scholar] [CrossRef] [Green Version]
- Hokama, M.; Oka, S.; Leon, J.; Ninomiya, T.; Honda, H.; Sasaki, K.; Iwaki, T.; Ohara, T.; Sasaki, T.; LaFerla, F.M.; et al. Altered expression of diabetes-related genes in Alzheimer’s disease brains: The Hisayama study. Cereb. Cortex 2014, 24, 2476–2488. [Google Scholar] [CrossRef]
- Durrenberger, P.F.; Fernando, F.S.; Kashefi, S.N.; Bonnert, T.P.; Seilhean, D.; Nait-Oumesmar, B.; Schmitt, A.; Gebicke-Haerter, P.J.; Falkai, P.; Grunblatt, E.; et al. Common mechanisms in neurodegeneration and neuroinflammation: A BrainNet Europe gene expression microarray study. J. Neural Transm. 2015, 122, 1055–1068. [Google Scholar] [CrossRef]
- Wang, M.; Roussos, P.; McKenzie, A.; Zhou, X.; Kajiwara, Y.; Brennand, K.J.; De Luca, G.C.; Crary, J.F.; Casaccia, P.; Buxbaum, J.D.; et al. Integrative network analysis of nineteen brain regions identifies molecular signatures and networks underlying selective regional vulnerability to Alzheimer’s disease. Genome Med. 2016, 8, 104. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.S.; Dunckley, T.; Beach, T.G.; Grover, A.; Mastroeni, D.; Walker, D.G.; Caselli, R.J.; Kukull, W.A.; McKeel, D.; Morris, J.C.; et al. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol. Genom. 2007, 28, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Berchtold, N.C.; Cribbs, D.H.; Coleman, P.D.; Rogers, J.; Head, E.; Kim, R.; Beach, T.; Miller, C.; Troncoso, J.; Trojanowski, J.Q.; et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc. Natl. Acad. Sci. USA 2008, 105, 15605–15610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berchtold, N.C.; Coleman, P.D.; Cribbs, D.H.; Rogers, J.; Gillen, D.L.; Cotman, C.W. Synaptic genes are extensively downregulated across multiple brain regions in normal human aging and Alzheimer’s disease. Neurobiol. Aging 2013, 34, 1653–1661. [Google Scholar] [CrossRef] [Green Version]
- Blalock, E.M.; Buechel, H.M.; Popovic, J.; Geddes, J.W.; Landfield, P.W. Microarray analyses of laser-captured hippocampus reveal distinct gray and white matter signatures associated with incipient Alzheimer’s disease. J. Chem. Neuroanat. 2011, 42, 118–126. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Cao, H.; Chen, J. False discovery rate control incorporating phylogenetic tree increases detection power in microbiome-wide multiple testing. Bioinformatics 2017, 33, 2873–2881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [Green Version]
- Mauri, E.; Sacchetti, A.; Vicario, N.; Peruzzotti-Jametti, L.; Rossi, F.; Pluchino, S. Evaluation of RGD functionalization in hybrid hydrogels as 3D neural stem cell culture systems. Biomater. Sci. 2018, 6, 501–510. [Google Scholar] [CrossRef]
- Box, G.E.; Tiao, G.C. Bayesian Inference in Statistical Analysis; John Wiley & Sons: Hoboken, NJ, USA, 1992. [Google Scholar] [CrossRef]
- Cheadle, C.; Vawter, M.P.; Freed, W.J.; Becker, K.G. Analysis of microarray data using Z score transformation. J. Mol. Diagn. 2003, 5, 73–81. [Google Scholar] [CrossRef] [Green Version]
- Scarpino, M.; Pinzone, M.R.; Di Rosa, M.; Madeddu, G.; Foca, E.; Martellotta, F.; Schioppa, O.; Ceccarelli, G.; Celesia, B.M.; d’Ettorre, G.; et al. Kidney disease in HIV-infected patients. Eur. Rev. Med. Pharmacol. Sci. 2013, 17, 2660–2667. [Google Scholar]
- Care, M.A.; Barrans, S.; Worrillow, L.; Jack, A.; Westhead, D.R.; Tooze, R.M. A microarray platform-independent classification tool for cell of origin class allows comparative analysis of gene expression in diffuse large B-cell lymphoma. PLoS ONE 2013, 8, e55895. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Coombes, K.R.; Highsmith, W.E.; Keating, M.J.; Abruzzo, L.V. Differences in gene expression between B-cell chronic lymphocytic leukemia and normal B cells: A meta-analysis of three microarray studies. Bioinformatics 2004, 20, 3166–3178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, T.B.; Riley, R.; Wymore, F.; Montgomery, P.; DeCaprio, D.; Engels, R.; Gellesch, M.; Hubble, J.; Jen, D.; Jin, H.; et al. TB database: An integrated platform for tuberculosis research. Nucleic Acids Res. 2009, 37, D499–D508. [Google Scholar] [CrossRef] [PubMed]
- Le Cao, K.A.; Rohart, F.; McHugh, L.; Korn, O.; Wells, C.A. YuGene: A simple approach to scale gene expression data derived from different platforms for integrated analyses. Genomics 2014, 103, 239–251. [Google Scholar] [CrossRef]
- Chen, Q.R.; Song, Y.K.; Wei, J.S.; Bilke, S.; Asgharzadeh, S.; Seeger, R.C.; Khan, J. An integrated cross-platform prognosis study on neuroblastoma patients. Genomics 2008, 92, 195–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasrebi, H.; Sperisen, P.; Praz, V.; Bucher, P. Can survival prediction be improved by merging gene expression data sets? PLoS ONE 2009, 4, e7431. [Google Scholar] [CrossRef] [Green Version]
- Mehmood, R.; El-Ashram, S.; Bie, R.; Dawood, H.; Kos, A. Clustering by fast search and merge of local density peaks for gene expression microarray data. Sci. Rep. 2017, 7, 45602. [Google Scholar] [CrossRef]
- Cheadle, C.; Cho-Chung, Y.S.; Becker, K.G.; Vawter, M.P. Application of z-score transformation to Affymetrix data. Appl. Bioinform. 2003, 2, 209–217. [Google Scholar]
- Feng, C.; Wu, J.; Yang, F.; Qiu, M.; Hu, S.; Guo, S.; Wu, J.; Ying, X.; Wang, J. Expression of Bcl-2 is a favorable prognostic biomarker in lung squamous cell carcinoma. Oncol. Lett. 2018, 15, 6925–6930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, C.; Huo, Y.; Xin, L.; Tian, B.; Yu, B. Feature selection and tumor classification for microarray data using relaxed Lasso and generalized multi-class support vector machine. J. Biol. 2019, 463, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Gulino, R.; Vicario, N.; Giunta, M.A.S.; Spoto, G.; Calabrese, G.; Vecchio, M.; Gulisano, M.; Leanza, G.; Parenti, R. Neuromuscular Plasticity in a Mouse Neurotoxic Model of Spinal Motoneuronal Loss. Int. J. Mol. Sci. 2019, 20, 1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vicario, N.; Bernstock, J.D.; Spitale, F.M.; Giallongo, C.; Giunta, M.A.S.; Li Volti, G.; Gulisano, M.; Leanza, G.; Tibullo, D.; Parenti, R.; et al. Clobetasol Modulates Adult Neural Stem Cell Growth via Canonical Hedgehog Pathway Activation. Int. J. Mol. Sci. 2019, 20, 1991. [Google Scholar] [CrossRef] [Green Version]
- Zetterberg, H.; Bozzetta, E.; Favole, A.; Corona, C.; Cavarretta, M.C.; Ingravalle, F.; Blennow, K.; Pocchiari, M.; Meloni, D. Neurofilaments in blood is a new promising preclinical biomarker for the screening of natural scrapie in sheep. PLoS ONE 2019, 14, e0226697. [Google Scholar] [CrossRef] [Green Version]
- Catrogiovanni, P.; Musumeci, G.; Giunta, S.; Imbesi, R.; Di Rosa, M. The expression levels of CHI3L1 and IL15Ralpha correlate with TGM2 in duodenum biopsies of patients with celiac disease. Inflamm. Res. 2020, 69, 925–935. [Google Scholar] [CrossRef]
- Musumeci, G.; Castrogiovanni, P.; Barbagallo, I.; Tibullo, D.; Sanfilippo, C.; Nunnari, G.; Pellicano, G.F.; Pavone, P.; Caltabiano, R.; Di Marco, R.; et al. Expression of the OAS Gene Family Is Highly Modulated in Subjects Affected by Juvenile Dermatomyositis, Resembling an Immune Response to a dsRNA Virus Infection. Int. J. Mol. Sci. 2018, 19, 2786. [Google Scholar] [CrossRef] [Green Version]
- Sanfilippo, C.; Pinzone, M.R.; Cambria, D.; Longo, A.; Palumbo, M.; Di Marco, R.; Condorelli, F.; Nunnari, G.; Malaguarnera, L.; Di Rosa, M. OAS Gene Family Expression Is Associated with HIV-Related Neurocognitive Disorders. Mol. Neurobiol. 2018, 55, 1905–1914. [Google Scholar] [CrossRef]
- Fagone, P.; Di Rosa, M.; Palumbo, M.; De Gregorio, C.; Nicoletti, F.; Malaguarnera, L. Modulation of heat shock proteins during macrophage differentiation. Inflamm. Res. 2012, 61, 1131–1139. [Google Scholar] [CrossRef] [PubMed]
- Caltabiano, R.; Castrogiovanni, P.; Barbagallo, I.; Ravalli, S.; Szychlinska, M.A.; Favilla, V.; Schiavo, L.; Imbesi, R.; Musumeci, G.; Di Rosa, M. Identification of Novel Markers of Prostate Cancer Progression, Potentially Modulated by Vitamin D. Appl. Sci. 2019, 9, 4923. [Google Scholar] [CrossRef] [Green Version]
- Nunnari, G.; Sanfilippo, C.; Castrogiovanni, P.; Imbesi, R.; Li Volti, G.; Barbagallo, I.; Musumeci, G.; Di Rosa, M. Network perturbation analysis in human bronchial epithelial cells following SARS-CoV2 infection. Exp. Cell Res. 2020, 395, 112204. [Google Scholar] [CrossRef]
- Sanfilippo, C.; Castrogiovanni, P.; Imbesi, R.; Tibullo, D.; Li Volti, G.; Barbagallo, I.; Vicario, N.; Musumeci, G.; Di Rosa, M. Middle-aged healthy women and Alzheimer’s disease patients present an overlapping of brain cell transcriptional profile. Neuroscience 2019, 406, 333–344. [Google Scholar] [CrossRef]
- Castrogiovanni, P.; Li Volti, G.; Sanfilippo, C.; Tibullo, D.; Galvano, F.; Vecchio, M.; Avola, R.; Barbagallo, I.; Malaguarnera, L.; Castorina, S.; et al. Fasting and Fast Food Diet Play an Opposite Role in Mice Brain Aging. Mol. Neurobiol. 2018, 55, 6881–6893. [Google Scholar] [CrossRef]
- D’Andrea, F.; Rullo, E.V.; Facciola, A.; Di Rosa, M.; Condorelli, F.; Marino, A.; Cacopardo, B.; Pellicano, G.F.; Nunnari, G.; Ceccarelli, M. Epstein Barr Virus related cancer in people living with HIV: A review of the literature. World Cancer Res. J. 2020, 7, e1512. [Google Scholar]
- Giallongo, S.; Di Rosa, M.; Caltabiano, R.; Longhitano, L.; Reibaldi, M.; Distefano, A.; Lo Re, O.; Amorini, A.M.; Puzzo, L.; Salvatorelli, L.; et al. Loss of macroH2A1 decreases mitochondrial metabolism and reduces the aggressiveness of uveal melanoma cells. Aging 2020, 12, 9745–9760. [Google Scholar] [CrossRef] [PubMed]
- Malaguarnera, L.; Nunnari, G.; Di Rosa, M. Nuclear import sequence identification in hOAS3 protein. Inflamm. Res. 2016, 65, 895–904. [Google Scholar] [CrossRef]
- Tibullo, D.; Di Rosa, M.; Giallongo, C.; La Cava, P.; Parrinello, N.L.; Romano, A.; Conticello, C.; Brundo, M.V.; Saccone, S.; Malaguarnera, L.; et al. Bortezomib modulates CHIT1 and YKL40 in monocyte-derived osteoclast and in myeloma cells. Front. Pharmacol. 2015, 6, 226. [Google Scholar] [CrossRef] [PubMed]
- Szychlinska, M.A.; Trovato, F.M.; Di Rosa, M.; Malaguarnera, L.; Puzzo, L.; Leonardi, R.; Castrogiovanni, P.; Musumeci, G. Co-Expression and Co-Localization of Cartilage Glycoproteins CHI3L1 and Lubricin in Osteoarthritic Cartilage: Morphological, Immunohistochemical and Gene Expression Profiles. Int. J. Mol. Sci. 2016, 17, 359. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Wang, N.; Riabov, V.; Mossel, D.M.; Larionova, I.; Schledzewski, K.; Trofimova, O.; Sevastyanova, T.; Zajakina, A.; Schmuttermaier, C.; et al. SI-CLP inhibits the growth of mouse mammary adenocarcinoma by preventing recruitment of tumor-associated macrophages. Int. J. Cancer 2020, 146, 1396–1408. [Google Scholar] [CrossRef] [Green Version]
- Penzes, P.; Vanleeuwen, J.E. Impaired regulation of synaptic actin cytoskeleton in Alzheimer’s disease. Brain Res. Rev. 2011, 67, 184–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mack, T.G.; Kreis, P.; Eickholt, B.J. Defective actin dynamics in dendritic spines: Cause or consequence of age-induced cognitive decline? Biol. Chem. 2016, 397, 223–229. [Google Scholar] [CrossRef]
- Gao, Y.; Tatavarty, V.; Korza, G.; Levin, M.K.; Carson, J.H. Multiplexed dendritic targeting of alpha calcium calmodulin-dependent protein kinase II, neurogranin, and activity-regulated cytoskeleton-associated protein RNAs by the A2 pathway. Mol. Biol. Cell 2008, 19, 2311–2327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorsell, A.; Bjerke, M.; Gobom, J.; Brunhage, E.; Vanmechelen, E.; Andreasen, N.; Hansson, O.; Minthon, L.; Zetterberg, H.; Blennow, K. Neurogranin in cerebrospinal fluid as a marker of synaptic degeneration in Alzheimer’s disease. Brain Res. 2010, 1362, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Sanfilippo, C.; Castrogiovanni, P.; Imbesi, R.; Di Rosa, M. CHI3L2 Expression Levels Are Correlated with AIF1, PECAM1, and CALB1 in the Brains of Alzheimer’s Disease Patients. J. Mol. Neurosci. 2020, 70, 1598–1610. [Google Scholar] [CrossRef] [PubMed]
- Di Rosa, M.; Dell’Ombra, N.; Zambito, A.M.; Malaguarnera, M.; Nicoletti, F.; Malaguarnera, L. Chitotriosidase and inflammatory mediator levels in Alzheimer’s disease and cerebrovascular dementia. Eur. J. Neurosci. 2006, 23, 2648–2656. [Google Scholar] [CrossRef]








| N° | Dataset | Organism | Platform | NDHC | AD | References | |
|---|---|---|---|---|---|---|---|
| 1 | GSE33000 | Homo sapiens | GPL4372 | HBTRC | 144 | 310 | [51] |
| 2 | GSE28894 | Homo sapiens | GPL6104 | NIA | 29 | 0 | [52] |
| 3 | GSE35978 | Homo sapiens | GPL6244 | SMRI | 13 | 0 | [53] |
| 4 | GSE15745 | Homo sapiens | GPL6104 | UMBB | 132 | 0 | [54] |
| 5 | GSE44772 | Homo sapiens | GPL4372 | HBTRC | 273 | 387 | [55] |
| 6 | GSE36192 | Homo sapiens | GPL6947 | NIA | 379 | 0 | [56] |
| 7 | GSE60862 | Homo sapiens | GPL5175 | UKBEC | 226 | 0 | [57] |
| 8 | GSE118553 | Homo sapiens | GPL10558 | MRC-LBB | 72 | 167 | [58] |
| 9 | GSE25219 | Homo sapiens | GPL5175 | BTBDDUM | 27 | 0 | [59] |
| 10 | GSE71620 | Homo sapiens | GPL11532 | PBTDP | 208 | 0 | [60] |
| 11 | GSE5392 | Homo sapiens | GPL96 | SMRIC | 3 | 0 | [61] |
| 12 | GSE36980 | Homo sapiens | GPL6244 | KU | 47 | 32 | [62] |
| 13 | GSE26927 | Homo sapiens | GPL6255 | BNEN | 4 | 11 | [63] |
| 14 | GSE84422 | Homo sapiens | GPL570 | MSBB | 28 | 74 | [64] |
| 15 | GSE5281 | Homo sapiens | GPL570 | ADCs | 74 | 87 | [65] |
| 16 | GSE48350 | Homo sapiens | GPL570 | ADC | 93 | 80 | [66] |
| 17 | GSE11882 | Homo sapiens | GPL570 | MSBB | 93 | 0 | [67] |
| 18 | GSE28146 | Homo sapiens | GPL570 | BBADRCUK | 8 | 22 | [68] |
| Age Stage | NDHCS | AD | |
|---|---|---|---|
| 1 | 53–65 middle-age | 734 = 582 male + 152 female | 59 = 34 male + 25 female |
| 2 | 65–75 senior | 425 = 291 male + 134 female | 219 = 75 male + 144 female |
| 3 | 76–89 elderly | 493 = 328 male + 165 female | 639 = 361 male + 278 female |
| 4 | 90–99 nonagenarian | 179 = 61 male + 118 female | 235 = 187 male + 48 female |
| 5 | >100 centenarian | 22 = 4 male + 18 female | 18 = 13 male + 5 female |
| Total sample | 1853 | 1170 |
| N° | Brain Regions | Abbreviations | Brain Portions | n° of Sample | Age |
|---|---|---|---|---|---|
| 1 | prefrontal | PFC | prefrontal cortex; dorsolateral prefrontal cortex; dorsolateral prefrontal cortex; medial prefrontal cortex; ofc (orbitofrontal cortex); orbitofrontal cortex; orbital prefrontal cortex; ventral forebrain; ventrolateral cortex; ventrolateral prefrontal cortex | 1045 = 784 male + 261 female | 51.18 ± 21 |
| 2 | frontal | FC | frontal cortex frontal pole (Brodmann area 9, 10); medial frontal cortex | 868 = 577 male + 291 female | 51.07 ± 26.08 |
| 3 | occipital | OC | occipital cortex; primary visual cortex; visual cortex | 286 = 207 male + 79 female | 55.16 ± 25.44 |
| 4 | cerebellum | CB | cerebellar cortex; cerebellum; upper (rostral) rhombic lip | 1009 = 693 male + 316 female | 50.9 ± 24.52 |
| 5 | temporal | TP | inferior temporal cortex; primary auditory cortex; superior temporal cortex; superior temporal cortex (Brodmann area 22); temporal cortex; ventral head of the caudate nucleus | 466 = 174 male + 292 female | 47.77 ± 29.01 |
| 6 | cingulate | CYN | anterior cingulate; caudal ganglionic eminence; lateral ganglionic eminence; medial ganglionic eminence; medial temporal gyrus; postcentral gyrus; posterior cingulate; posterior cingulate cortex; subpial grey matter lesions from the frontal gyri; superior frontal gyrus | 319 = 196 male + 123 female | 61.35 ± 22.55 |
| 7 | diencephalon | DIE | basal ganglia; dorsal thalamus; putamen; striatum; nucleus accumbens; substantia nigra; thalamus; mediodorsal nucleus of the thalamus | 450 = 304 male + 146 female | 55.41 ± 24.47 |
| 8 | limbic system | LS | amygdala; entorhinal cortex; hippocampus | 563 = 356 male + 207 female | 56.83 ± 26.31 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castrogiovanni, P.; Sanfilippo, C.; Imbesi, R.; Maugeri, G.; Lo Furno, D.; Tibullo, D.; Castorina, A.; Musumeci, G.; Di Rosa, M. Brain CHID1 Expression Correlates with NRGN and CALB1 in Healthy Subjects and AD Patients. Cells 2021, 10, 882. https://doi.org/10.3390/cells10040882
Castrogiovanni P, Sanfilippo C, Imbesi R, Maugeri G, Lo Furno D, Tibullo D, Castorina A, Musumeci G, Di Rosa M. Brain CHID1 Expression Correlates with NRGN and CALB1 in Healthy Subjects and AD Patients. Cells. 2021; 10(4):882. https://doi.org/10.3390/cells10040882
Chicago/Turabian StyleCastrogiovanni, Paola, Cristina Sanfilippo, Rosa Imbesi, Grazia Maugeri, Debora Lo Furno, Daniele Tibullo, Alessandro Castorina, Giuseppe Musumeci, and Michelino Di Rosa. 2021. "Brain CHID1 Expression Correlates with NRGN and CALB1 in Healthy Subjects and AD Patients" Cells 10, no. 4: 882. https://doi.org/10.3390/cells10040882