Proteomic Analysis of Ginseng (Panax ginseng C. A. Meyer) Fluid Proteins under Salt Stress
Abstract
:1. Introduction
2. Materials and Methods
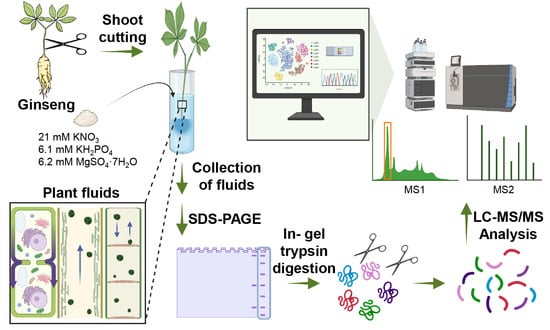
2.1. Plant Materials and Protein Extraction Using Ginseng Fluids and Leaves
2.2. Validation of Physiological Changes Using Ginseng Leaves and Culture Fluids and Western Blot
2.3. Shotgun Proteomic Analysis and Data Processing
2.4. Validation with Quantitative Real-Time PCR (qRT-PCR)
3. Results and Discussion
3.1. Salt-Stress-Triggered Accumulation of Reactive Oxygen Species in Ginseng
3.2. Extraction of Fluid Protein from Ginseng and SDS-PAGE Analysis
3.3. Application of Shotgun Proteomics for Identification of Salt-Responsive Proteins
3.4. Inhibition of Photosynthesis by Salt Stress in Ginseng
3.5. Accumulation of Energy Metabolism Related Proteins during Salt Stress
3.6. Activation of ROS Scavenging Enzymes by Oxidative Stress
3.7. Identification of Proteins Related to Various Metabolism
3.8. Validation of Identified Proteins Using qRT-PCR Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kumar, H.; Song, S.-Y.; More, S.V.; Kang, S.-M.; Kim, B.-W.; Kim, I.-S.; Choi, D.-K. Traditional korean east asian medicines and herbal formulations for cognitive impairment. Molecules 2013, 18, 14670–14693. [Google Scholar] [CrossRef] [PubMed]
- Sofowora, A.; Ogunbodede, E.; Onayade, A. The role and place of medicinal plants in the strategies for disease prevention. Afr. J. Tradit. Complement. Altern. Med. 2013, 10, 210–229. [Google Scholar] [CrossRef] [PubMed]
- Ratan, Z.A.; Haidere, M.F.; Hong, Y.H.; Park, S.H.; Lee, J.-O.; Lee, J.; Cho, J.Y. Pharmacological potential of ginseng and its major component ginsenosides. J. Ginseng Res. 2021, 45, 199–210. [Google Scholar] [CrossRef]
- Lee, C.H.; Kim, J.-H. A review on the medicinal potentials of ginseng and ginsenosides on cardiovascular siseases. J. Ginseng Res. 2014, 38, 161–166. [Google Scholar] [CrossRef]
- Cho, D.-H.; Park, K.-J.; Yu, Y.-H.; Oh, S.-H.; Lee, H.-S. Root-rot development of 2-year old ginseng (Panax ginseng C. A. Meyer) caused by Cylindrocarpon destructans (zinssm.) scholten in the continuous cultivation field. J. Ginseng Res. 1995, 19, 175–180. [Google Scholar]
- Kim, Y.-S.; Park, C.-S.; Lee, D.-Y.; Lee, J.-S.; Lee, S.-H.; In, J.-G.; Hong, T.-K. Phenological growth stages of korean ginseng (Panax ginseng) according to the extended BBCH scale. J. Ginseng Res. 2021, 45, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Lee, O.; Sathiyaraj, G.; Kim, Y.-J.; In, J.-G.; Kwon, W.-S.; Kim, J.-H.; Yang, D.-C. Defense genes induced by pathogens and abiotic stresses in Panax ginseng C. A. Meyer. J. Ginseng Res. 2011, 35, 1–11. [Google Scholar] [CrossRef]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant salinity stress: Many unanswered questions remain. Front. Plant Sci. 2019, 10, 80. [Google Scholar] [CrossRef]
- Onyekachi, O.G.; Boniface, O.O.; Gemlack, N.F.; Nicholas, N. The effect of climate change on abiotic plant stress: A review. In Biotic and Abiotic Stresses in Plants; Oliverira, A.D., Ed.; IntechOpen: London, UK, 2019; p. 17. [Google Scholar] [CrossRef]
- Howard, R.J.; Mendelssohn, I. A salinity as a constraint on growth of oligohaline marsh macrophytes species variation in stress tolerance. Am. J. Bot. 1999, 86, 785–794. [Google Scholar] [CrossRef]
- Riyazuddin, R.; Verma, R.; Singh, K.; Nisha, N.; Keisham, M.; Bhati, K.K.; Kim, S.T.; Gupta, R. Ethylene: A master regulator of salinity stress tolerance in plants. Biomolules 2020, 10, 959. [Google Scholar] [CrossRef]
- Kim, S.T.; Bae, D.-W.; Lee, K.; Hwang, J.E.; Bang, K.-H.; Kim, Y.-C.; Kim, O.-T.; Yoo, N.H.; Kang, K.Y.; Hyun, D.Y.; et al. Proteomic analysis of korean ginseng (Panax ginseng C. A. Meyer) following exposure to salt stress. J. Plant Biotechnol. 2008, 35, 185–193. [Google Scholar] [CrossRef]
- Kim, S.W.; Min, C.W.; Gupta, R.; Jo, I.H.; Bang, K.H.; Kim, Y.-C.; Kim, K.-H.; Kim, S.T. Proteomics analysis of early salt-responsive proteins in ginseng (Panax Ginseng C. A. Meyer) leaves. Korean J. Med. Crop Sci. 2014, 22, 398–404. [Google Scholar] [CrossRef]
- Rodríguez-Celma, J.; Ceballos-Laita, L.; Grusak, M.A.; Abadía, J.; López-Millán, A.-F. Plant fluid proteomics: Delving into the xylem sap, phloem sap and apoplastic fluid proteomes. Biochim. Biophys. Acta-Proteins Proteomics 2016, 1864, 991–1002. [Google Scholar] [CrossRef] [PubMed]
- Carella, P.; Wilson, D.C.; Kempthorne, C.J.; Cameron, R.K. Vascular sap proteomics: Providing insight into long-distance signaling during stress. Front. Plant Sci. 2016, 7, 651. [Google Scholar] [CrossRef] [PubMed]
- Sattelmacher, B.; Mühling, K.H.; Pennewiß, K. The apoplast-Its significance for the nutrition of higher plants. J. Plant Nutr. Soil. Sci. 1998, 161, 485–498. [Google Scholar] [CrossRef]
- Jorrin-Novo, J.V. Plant proteomics methods and protocols. In Plant Proteomics: Methods and Protocols, 2nd ed.; Jorrin-Novo, J.V., Komatsu, S., Weckwerth, W., Wienkoop, S., Eds.; Human Press: Totowa, NJ, USA, 2014; Volume 1072, pp. 3–13. [Google Scholar] [CrossRef]
- Rep, M.; Dekker, H.L.; Vossen, J.H.; de Boer, A.D.; Houterman, P.M.; Speijer, D.; Back, J.W.; de Koster, C.G.; Cornelissen, B.J.C. Mass spectrometric identification of isoforms of PR proteins in xylem sap of fungus-infected tomato. Plant Physiol. 2002, 130, 904–917. [Google Scholar] [CrossRef]
- Gawehns, F.; Ma, L.; Bruning, O.; Houterman, P.M.; Boeren, S.; Cornelissen, B.J.C.; Rep, M.; Takken, F.L.W. The effector repertoire of Fusarium oxysporum determines the tomato xylem proteome composition following infection. Front. Plant Sci. 2015, 6, 967. [Google Scholar] [CrossRef]
- Pu, Z.; Ino, Y.; Kimura, Y.; Tago, A.; Shimizu, M.; Natsume, S.; Sano, Y.; Fujimoto, R.; Kaneko, K.; Shea, D.J.; et al. Changes in the proteome of xylem sap in Brassica oleracea in response to Fusarium oxysporum stress. Front. Plant Sci. 2016, 7, 31. [Google Scholar] [CrossRef]
- Abeysekara, N.S.; Bhattacharyya, M.K. Analyses of the xylem sap proteomes identified candidate Fusarium virguliforme proteinacious toxins. PLoS ONE 2014, 9, e93667. [Google Scholar] [CrossRef] [Green Version]
- Figueiredo, J.; Cavaco, A.R.; Guerra-Guimarães, L.; Leclercq, C.; Renaut, J.; Cunha, J.; Eiras-Dias, J.; Cordeiro, C.; Matos, A.R.; Sousa Silva, M.; et al. An apoplastic fluid extraction method for the characterization of grapevine leaves proteome and metabolome from a single sample. Physiol. Plant. 2021, 171, 343–357. [Google Scholar] [CrossRef]
- Fernandez-Garcia, N.; Hernandez, M.; Casado-Vela, J.; Bru, R.; Elortza, F.; Hedden, P.; Olmos, E. Changes to the proteome and targeted metabolites of xylem sap in Brassica oleracea in response to salt stress. Plant. Cell Environ. 2011, 34, 821–836. [Google Scholar] [CrossRef]
- Nedbal, L.; Soukupová, J.; Kaftan, D.; Whitmarsh, J.; Trtílek, M. Kinetic imaging of chlorophyll fluorescence using modulated light. Photosynth. Res. 2000, 66, 3–12. [Google Scholar] [CrossRef]
- Meng, Q.; Gupta, R.; Min, C.W.; Kim, J.; Kramer, K.; Wang, Y.; Park, S.R.; Finkemeier, I.; Kim, S.T. A proteomic insight into the MSP1 and flg22 induced signaling in Oryza sativa leaves. J. Proteom. 2019, 196, 120–130. [Google Scholar] [CrossRef]
- Kim, S.T.; Wang, Y.; Kang, S.Y.; Kim, S.G.; Rakwal, R.; Kim, Y.C.; Kang, K.Y. Developing rice embryo proteomics reveals essential role for embryonic proteins in regulation of seed germination. J. Proteome Res. 2009, 8, 3598–3605. [Google Scholar] [CrossRef]
- Kim, Y.J.; Wang, Y.; Gupta, R.; Kim, S.W.; Min, C.W.; Kim, Y.C.; Park, K.H.; Agrawal, G.K.; Rakwal, R.; Choung, M.G.; et al. Protamine sulfate precipitation method depletes abundant plant seed-storage proteins: A case study on legume plants. Proteomics 2015, 15, 1760–1764. [Google Scholar] [CrossRef]
- Gupta, R.; Lee, S.J.; Min, C.W.; Kim, S.W.; Park, K.-H.; Bae, D.-W.; Lee, B.W.; Agrawal, G.K.; Rakwal, R.; Kim, S.T. Coupling of gel-based 2-DE and 1-DE shotgun proteomics approaches to dig deep into the leaf senescence proteome of Glycine max. J. Proteom. 2016, 148, 65–74. [Google Scholar] [CrossRef]
- Gupta, R.; Min, C.W.; Kim, S.W.; Wang, Y.; Agrawal, G.K.; Rakwal, R.; Kim, S.G.; Lee, B.W.; Ko, J.M.; Baek, I.Y.; et al. Comparative investigation of seed coats of brown- versus yellow-colored soybean seeds using an integrated proteomics and metabolomics approach. Proteomics 2015, 15, 1706–1716. [Google Scholar] [CrossRef]
- Kim, S.W.; Gupta, R.; Lee, S.H.; Min, C.W.; Agrawal, G.K.; Rakwal, R.; Kim, J.B.; Jo, I.H.; Park, S.-Y.; Kim, J.K.; et al. An integrated biochemical, proteomics, and metabolomics approach for supporting medicinal value of Panax ginseng fruits. Front. Plant Sci. 2016, 7, 994. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Jo, I.H.; Lee, J.; Hong, C.E.; Lee, D.J.; Bae, W.; Park, S.G.; Ahn, Y.J.; Kim, Y.C.; Kim, J.U.; Lee, J.W.; et al. Isoform sequencing provides a more comprehensive view of the Panax ginseng transcriptome. Genes 2017, 8, 228. [Google Scholar] [CrossRef]
- Wang, K.; Jiang, S.; Sun, C.; Lin, Y.; Yin, R.; Wang, Y.; Zhang, M. The spatial and temporal transcriptomic landscapes of ginseng, Panax ginseng C. A. Meyer. Sci. Rep. 2016, 5, 18283. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Csordas, A.; Noemi, D.-T.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Yasset, P.-R.; Reisinger, F.; Ternent, T. 2016 Update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. AgriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 Update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]
- Usadel, B.; Poree, F.; Nagel, A.; Lohse, M.; Czedik-Eysenberg, A.; Stitt, M. A guide to using MapMan to visualize and compare omics data in plants: A case study in the crop species, maize. Plant Cell Environ. 2009, 32, 1211–1229. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Salt stress signaling and mechanisms of plant salt tolerance. In Genetic Engineering: Principles and Methods; Setlow, J.K., Ed.; Springer Nature: Berlin, Germany, 2006; pp. 141–177. [Google Scholar] [CrossRef]
- Lee, D.H.; Kim, Y.S.; Lee, C.B. The inductive responses of the antioxidant enzymes by salt stress in the rice (Oryza Sativa L.). J. Plant Physiol. 2001, 158, 737–745. [Google Scholar] [CrossRef]
- Quick, W.P.; Horton, P.; Walker, D.A. Studies on the induction of chlorophyll fluorescence in barley protoplasts. I. factors affecting the observation of oscillations in the yield of chlorophyll fluorescence and the rate of oxygen evolution. Proc. R. Soc. Lond. Ser. B. Biol. Sci. 1984, 220, 361–370. [Google Scholar] [CrossRef]
- Berwal, M.K.; Ram, C. Superoxide dismutase: A stable biochemical marker for abiotic stress tolerance in higher plants. In Biotic and Abiotic Stresses in Plants; Oliverira, A.D., Ed.; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef]
- Ligat, L.; Lauber, E.; Albenne, C.; Clemente, H.S.; Valot, B.; Zivy, M.; Pont-Lezica, R.; Arlat, M.; Jamet, E. Analysis of the xylem sap proteome of Brassica oleracea reveals a high content in secreted proteins. Proteomics 2011, 11, 1798–1813. [Google Scholar] [CrossRef] [Green Version]
- Djordjevic, M.A.; Oakes, M.; Li, D.X.; Hwang, C.H.; Hocart, C.H.; Gresshoff, P.M. The Glycine max xylem sap and apoplast proteome. J. Proteome Res. 2007, 6, 3771–3779. [Google Scholar] [CrossRef]
- Bahrun, A.; Jensen, C.R.; Asch, F.; Mogensen, V.O. Drought-induced changes in xylem pH, ionic composition, and ABA concentration act as early signals in field-grown maize (Zea mays L.). J. Exp. Bot. 2002, 53, 251–263. [Google Scholar] [CrossRef]
- Tetyuk, O.; Benning, U.F.; Hoffmann-Benning, S. Collection and analysis of arabidopsis phloem exudates using the EDTA-facilitated method. JoVE 2013, 80, e51111. [Google Scholar] [CrossRef]
- Tanaka, A.; Makino, A. Photosynthetic research in plant science. Plant Cell Physiol. 2009, 50, 681–683. [Google Scholar] [CrossRef]
- Sobhanian, H.; Aghaei, K.; Komatsu, S. Changes in the plant proteome resulting from salt stress: Toward the creation of salt-tolerant crops? J. Proteom. 2011, 74, 1323–1337. [Google Scholar] [CrossRef] [PubMed]
- Helena, H.; František, H.; Jaroslava, M.; Kamil, K. Effects of salt stress on water status, photosynthesis and chlorophyll fluorescence of rocket. Plant Soil Environ. 2017, 63, 362–367. [Google Scholar] [CrossRef]
- Xiong, J.; Sun, Y.; Yang, Q.; Tian, H.; Zhang, H.; Liu, Y.; Chen, M. Proteomic analysis of early salt stress responsive proteins in alfalfa roots and shoots. Proteome Sci. 2017, 15, 19. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Luo, Q.; Wang, Q.; Zhang, X.; Qi, Z.; Xu, F.; Lei, X.; Cao, Y.; Chow, W.S.; Sun, G. Physiological and proteomic responses to salt stress in chloroplasts of diploid and tetraploid black locust (Robinia pseudoacacia L.). Sci. Rep. 2016, 6, 23098. [Google Scholar] [CrossRef]
- Lu, W.; Tang, X.; Huo, Y.; Xu, R.; Qi, S.; Huang, J.; Zheng, C.; Wu, C. Identification and characterization of fructose 1,6-bisphosphate aldolase genes in arabidopsis reveal a gene family with diverse responses to abiotic stresses. Gene 2012, 503, 65–74. [Google Scholar] [CrossRef]
- Lydakis-Simantiris, N.; Hutchison, R.S.; Betts, S.D.; Barry, B.A.; Yocum, C.F. Manganese stabilizing protein of photosystem II is a thermostable, natively unfolded polypeptide. Biochemistry 1999, 38, 404–414. [Google Scholar] [CrossRef]
- Lundin, B.; Hansson, M.; Schoefs, B.; Vener, A.V.; Spetea, C. The arabidopsis PsbO2 protein regulates dephosphorylation and turnover of the photosystem ii reaction centre D1 protein. Plant J. 2007, 49, 528–539. [Google Scholar] [CrossRef]
- Ettinger, W.F.; Theg, S.M. Physiologically active chloroplasts contain pools of unassembled extrinsic proteins of the photosynthetic oxygen-evolving enzyme complex in the thylakoid lumen. J. Cell Biol. 1991, 115, 321–328. [Google Scholar] [CrossRef]
- Razavizadeh, R.; Ehsanpour, A.A.; Ahsan, N.; Komatsu, S. Proteome analysis of tobacco leaves under salt stress. Peptides 2009, 30, 1651–1659. [Google Scholar] [CrossRef]
- Givan, C.V. Evolving concepts in plant glycolysis: Two centuries of progress. Biol. Rev. 1999, 74, 277–309. [Google Scholar] [CrossRef]
- Li, C.; Sun, J.; Qi, X.; Liu, L. NaCl stress impact on the key enzymes in glycolysis from Lactobacillus bulgaricus during freeze-drying. Braz. J. Microbiol. 2015, 46, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Sobhanian, H.; Motamed, N.; Jazii, F.R.; Nakamura, T.; Komatsu, S. Salt stress induced differential proteome and metabolome response in the shoots of Aeluropus lagopoides (poaceae), a halophyte C4 plant. J. Proteome Res. 2010, 9, 2882–2897. [Google Scholar] [CrossRef] [PubMed]
- Chitteti, B.R.; Peng, Z. Proteome and phosphoproteome differential expression under salinity stress in rice (Oryza Sativa) Roots. J. Proteome Res. 2007, 6, 1718–1727. [Google Scholar] [CrossRef]
- Sobhanian, H.; Razavizadeh, R.; Nanjo, Y.; Ehsanpour, A.; Jazii, F.; Motamed, N.; Komatsu, S. Proteome analysis of soybean leaves, hypocotyls and roots under salt stress. Proteome Sci. 2010, 8, 19. [Google Scholar] [CrossRef]
- Kaur, C.; Singla-Pareek, S.L.; Sopory, S.K. Glyoxalase and methylglyoxal as biomarkers for plant stress tolerance. Crit. Rev. Plant Sci. 2014, 33, 429–456. [Google Scholar] [CrossRef]
- Gossett, D.R.; Milhollon, E.P.; Lucas, M.C. Antioxidant response to NaCl stress in salt-tolerant and salt sensitive cultivars of cotton. Crop Sci. 1994, 34, 706–714. [Google Scholar] [CrossRef]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ros-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 53. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Anee, T.I.; Parvin, K.; Nahar, K.; Mahmud, J.A.; Fujita, M. Regulation of ascorbate-glutathione pathway in mitigating oxidative damage in plants under abiotic stress. Antioxidants 2019, 8, 384. [Google Scholar] [CrossRef]
- Smirnoff, N.; Arnaud, D. Hydrogen peroxide metabolism and functions in plants. New Phytol. 2019, 221, 1197–1214. [Google Scholar] [CrossRef]
- Yoshimura, K.; Miyao, K.; Gaber, A.; Takeda, T.; Kanaboshi, H.; Miyasaka, H.; Shigeoka, S. Enhancement of stress tolerance in transgenic tobacco plants overexpressing chlamydomonas glutathione peroxidase in chloroplasts or cytosol. Plant J. 2004, 37, 21–33. [Google Scholar] [CrossRef]
- Meloni, D.A.; Oliva, M.A.; Martinez, C.A.; Cambraia, J. Photosynthesis and activity of superoxide dismutase, peroxidase and glutathione reductase in cotton under salt stress. Environ. Exp. Bot. 2003, 49, 69–76. [Google Scholar] [CrossRef]
- Park, C.-J.; Seo, Y.-S. Heat shock proteins: A review of the molecular chaperones for plant immunity. plant Pathol. J. 2015, 31, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Mogk, A.; Kummer, E.; Bukau, B. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front. Mol. Biosci. 2015, 2, 22. [Google Scholar] [CrossRef] [PubMed]
- Parcerisa, I.L.; Rosano, G.L.; Ceccarelli, E.A. Biochemical characterization of ClpB3, a Chloroplastic disaggregase from Arabidopsis thaliana. Plant Mol. Biol. 2020, 104, 451–465. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Minic, Z. Physiological roles of plant glycoside hydrolases. Planta 2007, 227, 723. [Google Scholar] [CrossRef]
- Leubner-Metzger, G. Functions and regulation of β-1,3-glucanases during seed germination, dormancy release and after-ripening. Seed Sci. Res. 2003, 2003 13, 17–34. [Google Scholar] [CrossRef]
- Cao, Y.-Y.; Yang, J.-F.; Liu, T.-Y.; Su, Z.-F.; Zhu, F.-Y.; Chen, M.-X.; Fan, T.; Ye, N.-H.; Feng, Z.; Wang, L.-J.; et al. A phylogenetically informed comparison of GH1 hydrolases between arabidopsis and rice response to stressors. Front. Plant Sci. 2017, 8, 350. [Google Scholar] [CrossRef]
- Gottschalk, M.; Dolgener, E.; Xoconostle-Cázares, B.; Lucas, W.J.; Komor, E.; Schobert, C. Ricinuscommunis cyclophilin: Functional characterisation of a sieve tube protein involved in protein folding. Planta 2008, 228, 687. [Google Scholar] [CrossRef]
- Aigen, F.; Zengyong, H.; Sun, C.H.; Amparo, L.; Buchanan, B.B.; Sheng, L. A chloroplast cyclophilin functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2007, 104, 15947–15952. [Google Scholar] [CrossRef]
- Kang, B.; Zhang, Z.; Wang, L.; Zheng, L.; Mao, W.; Li, M.; Wu, Y.; Wu, P.; Mo, X. OsCYP2, a chaperone involved in degradation of auxin-responsive proteins, plays crucial roles in rice lateral root initiation. Plant J. 2013, 74, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Shapiguzov, A.; Edvardsson, A.; Vener, A.V. Profound redox sensitivity of peptidyl-prolyl isomerase activity in Arabidopsis thylakoid lumen. FEBS Lett. 2006, 580, 3671–3676. [Google Scholar] [CrossRef]
- Sekhar, K.; Priyanka, B.; Reddy, V.D.; Rao, K.V. Isolation and characterization of a pigeonpea cyclophilin (CcCYP) gene, and its over-expression in arabidopsis confers multiple abiotic stress tolerance. Plant. Cell Environ. 2010, 33, 1324–1338. [Google Scholar] [CrossRef]
- Min, C.W.; Lee, S.H.; Cheon, Y.E.; Han, W.Y.; Ko, J.M.; Kang, H.W.; Kim, Y.C.; Agrawal, G.K.; Rakwal, R.; Gupta, R.; et al. In-depth proteomic analysis of Glycine max seeds during controlled deterioration treatment reveals a shift in seed metabolism. J. Proteomics 2017, 169, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Alavilli, H.; Lee, H.; Park, M.; Yun, D.-J.; Lee, B. Enhanced multiple stress tolerance in Arabidopsis by overexpression of the polar moss peptidyl prolyl isomerase FKBP12 gene. Plant Cell Rep. 2018, 37, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, L.-J.; Huang, R.-D. Cytoskeleton and plant salt stress tolerance. Plant Signal. Behav. 2011, 6, 29–31. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Zhang, L.; Yuan, M.; Ge, Y.; Liu, Y.; Fan, J.; Ruan, Y.; Cui, Z.; Tong, S.; Zhang, S. The microfilament cytoskeleton plays a vital role in salt and osmotic stress tolerance in Arabidopsis. Plant Biol. 2010, 12, 70–78. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, J.-Y.; Min, C.W.; Kim, S.W.; Gupta, R.; Jang, W.; Bang, K.-H.; Kim, Y.-J.; Jo, I.-H.; Kim, S.T. Proteomic Analysis of Ginseng (Panax ginseng C. A. Meyer) Fluid Proteins under Salt Stress. Agronomy 2022, 12, 2048. https://doi.org/10.3390/agronomy12092048
Jung J-Y, Min CW, Kim SW, Gupta R, Jang W, Bang K-H, Kim Y-J, Jo I-H, Kim ST. Proteomic Analysis of Ginseng (Panax ginseng C. A. Meyer) Fluid Proteins under Salt Stress. Agronomy. 2022; 12(9):2048. https://doi.org/10.3390/agronomy12092048
Chicago/Turabian StyleJung, Ju-Young, Cheol Woo Min, So Wun Kim, Ravi Gupta, Woojong Jang, Kyong-Hwan Bang, Yu-Jin Kim, Ick-Hyun Jo, and Sun Tae Kim. 2022. "Proteomic Analysis of Ginseng (Panax ginseng C. A. Meyer) Fluid Proteins under Salt Stress" Agronomy 12, no. 9: 2048. https://doi.org/10.3390/agronomy12092048