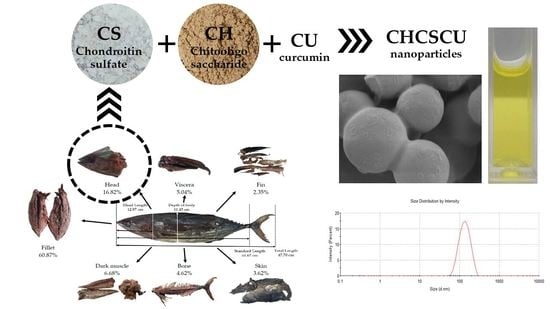
Nanoparticles Based on Chondroitin Sulfate from Tuna Heads and Chitooligosaccharides for Enhanced Water Solubility and Sustained Release of Curcumin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Measurement of Fish Length and Quantification of Components of Tuna-Processing By-Products
2.3. Optimization of Enzymatic Extraction of Chondroitin Sulfate from Tuna Heads
2.3.1. Sample Preparation
2.3.2. Optimization of Enzymatic Extraction of CS from Tuna Heads
2.4. Preparation and Characterization of Curcumin-Loaded Chondroitin Sulfate–Chitooligosaccharide (CHCSCU) Nanoparticles
2.4.1. Preparation of CU-Loaded Nanoparticles
2.4.2. Characterization of CU-Loaded Nanoparticles
2.4.3. In Vitro Release Profiles
2.4.4. Determination of Antioxidant Activities
2.5. Statistical Analysis
3. Results and Discussion
3.1. Measurement of Fish Length and Quantification of Components of Tuna-Processing By-Products
3.2. Optimization of Enzymatic Extraction of Chondroitin Sulfate from Tuna Heads
3.3. Effects of CH:CS Ratio on CU-Loaded Nanoparticle Characteristics
3.4. Characterization of Curcumin-Loaded Chitooligosaccharide–Chondroitin Sulfate (CHCSCU) Nanoparticles
3.4.1. FTIR Spectra
3.4.2. Nanoparticle Morphology
3.4.3. In Vitro Release Profiles
3.4.4. Antioxidant Activities
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abdallah, M.M.; Fernandez, N.; Matias, A.A.; Bronze, M.D.R. Hyaluronic acid and Chondroitin sulfate from marine and terrestrial sources: Extraction and purification methods. Carbohydr. Polym. 2020, 243, 116441. [Google Scholar] [CrossRef]
- Sharma, R.; Kuche, K.; Thakor, P.; Bhavana, V.; Srivastava, S.; Mehra, N.K.; Jain, S. Chondroitin Sulfate: Emerging biomaterial for biopharmaceutical purpose and tissue engineering. Carbohydr. Polym. 2022, 286, 119305. [Google Scholar] [CrossRef]
- Li, W.; Ura, K.; Takagi, Y. Industrial application of fish cartilaginous tissues. Curr. Res. Food Sci. 2022, 5, 698–709. [Google Scholar] [CrossRef]
- Urbi, Z.; Azmi, N.S.; Ming, L.C.; Hossain, M.S. A Concise Review of Extraction and Characterization of Chondroitin Sulphate from Fish and Fish Wastes for Pharmacological Application. Curr. Issues Mol. Biol. 2022, 44, 268. [Google Scholar] [CrossRef]
- Vazquez, J.A.; Fraguas, J.; Novoa-Carvallal, R.; Reis, R.L.; Antelo, L.T.; Perez-Martin, R.I.; Valcarcel, J. Isolation and Chemical Characterization of Chondroitin Sulfate from Cartilage By-Products of Blackmouth Catshark (Galeus melastomus). Mar. Drugs 2018, 16, 344. [Google Scholar] [CrossRef]
- Shionoya, K.; Suzuki, T.; Takada, M.; Sato, K.; Onishi, S.; Dohmae, N.; Nishino, K.; Wada, T.; Linhardt, R.J.; Toida, T.; et al. Comprehensive analysis of chondroitin sulfate and aggrecan in the head cartilage of bony fishes: Identification of proteoglycans in the head cartilage of sturgeon. Int. J. Biol. Macromol. 2022, 208, 333–342. [Google Scholar] [CrossRef]
- Vazquez, J.A.; Blanco, M.; Fraguas, J.; Pastrana, L.; Perez-Martin, R. Optimisation of the extraction and purification of chondroitin sulphate from head by-products of Prionace glauca by environmental friendly processes. Food Chem. 2016, 198, 28–35. [Google Scholar] [CrossRef]
- Blanco, M.; Fraguas, J.; Sotelo, C.G.; Perez-Martin, R.I.; Vazquez, J.A. Production of Chondroitin Sulphate from Head, Skeleton and Fins of Scyliorhinus canicula By-Products by Combination of Enzymatic, Chemical Precipitation and Ultrafiltration Methodologies. Mar. Drugs 2015, 13, 3287–3308. [Google Scholar] [CrossRef]
- Chuenban, P.; Sornsaruht, P.; Pimdee, P. How brand attitude, brand quality, and brand value affect Thai canned tuna consumer brand loyalty. Heliyon 2021, 7, e06301. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture 2022. Towards Blue Transformation; FAO: Rome, Italy, 2022. [Google Scholar] [CrossRef]
- Thai Tuna Industry Association. Thai Tuna Statistics of Import and Export in 2019–2021. Available online: https://thaituna.org/main/downloads/statistics/Update%20TH%20Export%20&%20Import_Tuna%20&%20Other%20fish_%202019%20-%202021.pdf (accessed on 30 August 2022).
- Kandyliari, A.; Mallouchos, A.; Papandroulakis, N.; Golla, J.P.; Lam, T.T.; Sakellari, A.; Karavoltsos, S.; Vasiliou, V.; Kapsokefalou, M. Nutrient Composition and Fatty Acid and Protein Profiles of Selected Fish By-Products. Foods 2020, 9, 190. [Google Scholar] [CrossRef]
- Rustad, T.; Storrø, I.; Slizyte, R. Possibilities for the utilisation of marine by-products. Int. J. Food Sci. Technol. 2011, 46, 2001–2014. [Google Scholar] [CrossRef]
- Coppola, D.; Lauritano, C.; Palma Esposito, F.; Riccio, G.; Rizzo, C.; de Pascale, D. Fish Waste: From Problem to Valuable Resource. Mar. Drugs 2021, 19, 116. [Google Scholar] [CrossRef]
- Gamarro, E.G.; Worawattanamateekul, W.; Sentina, J.; Gopal, S. By-products of tuna processing. Globefish Res. Progr. 2013, 112, 48. [Google Scholar]
- Wu, T.H.; Nigg, J.D.; Stine, J.J.; Bechtel, P.J. Nutritional and Chemical Composition of By-Product Fractions Produced from Wet Reduction of Individual Red Salmon (Oncorhynchus nerka) Heads and Viscera. J. Aquat. Food Prod. Technol. 2011, 20, 183–195. [Google Scholar] [CrossRef]
- Krishnamoorthy, S.; Paranthaman, R.; Moses, J.A.; Anandharamakrishnan, C. Curcumin. In Nutraceuticals and Health Care; Academic Press: Cambridge, MA, USA, 2022; pp. 159–175. [Google Scholar]
- Sohn, S.I.; Priya, A.; Balasubramaniam, B.; Muthuramalingam, P.; Sivasankar, C.; Selvaraj, A.; Valliammai, A.; Jothi, R.; Pandian, S. Biomedical Applications and Bioavailability of Curcumin-An Updated Overview. Pharmaceutics 2021, 13, 2102. [Google Scholar] [CrossRef]
- Tomeh, M.A.; Hadianamrei, R.; Zhao, X. A Review of Curcumin and Its Derivatives as Anticancer Agents. Int. J. Mol. Sci. 2019, 20, 1033. [Google Scholar] [CrossRef]
- He, Y.; Yue, Y.; Zheng, X.; Zhang, K.; Chen, S.; Du, Z. Curcumin, inflammation, and chronic diseases: How are they linked? Molecules 2015, 20, 9183–9213. [Google Scholar] [CrossRef]
- Nebrisi, E.E. Neuroprotective Activities of Curcumin in Parkinson's Disease: A Review of the Literature. Int. J. Mol. Sci. 2021, 22, 11248. [Google Scholar] [CrossRef]
- Jakubczyk, K.; Druzga, A.; Katarzyna, J.; Skonieczna-Zydecka, K. Antioxidant Potential of Curcumin-A Meta-Analysis of Randomized Clinical Trials. Antioxidants 2020, 9, 1092. [Google Scholar] [CrossRef]
- Mimica, B.; Bucevic Popovic, V.; Banjari, I.; Jelicic Kadic, A.; Puljak, L. Methods Used for Enhancing the Bioavailability of Oral Curcumin in Randomized Controlled Trials: A Meta-Research Study. Pharmaceuticals 2022, 15, 939. [Google Scholar] [CrossRef]
- Chen, Y.; Lu, Y.; Lee, R.J.; Xiang, G. Nano Encapsulated Curcumin: And Its Potential for Biomedical Applications. Int. J. Nanomed. 2020, 15, 3099–3120. [Google Scholar] [CrossRef]
- Suresh, K.; Nangia, A. Curcumin: Pharmaceutical solids as a platform to improve solubility and bioavailability. CrystEngComm 2018, 20, 3277–3296. [Google Scholar] [CrossRef]
- Park, H.R.; Rho, S.-J.; Kim, Y.-R. Solubility, stability, and bioaccessibility improvement of curcumin encapsulated using 4-α-glucanotransferase-modified rice starch with reversible pH-induced aggregation property. Food Hydrocoll. 2019, 95, 19–32. [Google Scholar] [CrossRef]
- Tabanelli, R.; Brogi, S.; Calderone, V. Improving Curcumin Bioavailability: Current Strategies and Future Perspectives. Pharmaceutics 2021, 13, 1715. [Google Scholar] [CrossRef]
- Chopra, H.; Dey, P.S.; Das, D.; Bhattacharya, T.; Shah, M.; Mubin, S.; Maishu, S.P.; Akter, R.; Rahman, M.H.; Karthika, C.; et al. Curcumin Nanoparticles as Promising Therapeutic Agents for Drug Targets. Molecules 2021, 26, 4998. [Google Scholar] [CrossRef]
- Hussain, S.; Ashafaq, M.; Alshahrani, S.; Bokar, I.A.M.; Siddiqui, R.; Alam, M.I.; Taha, M.M.E.; Almoshari, Y.; Alqahtani, S.S.; Ahmed, R.A.; et al. Hepatoprotective Effect of Curcumin Nano-Lipid Carrier against Cypermethrin Toxicity by Countering the Oxidative, Inflammatory, and Apoptotic Changes in Wistar Rats. Molecules 2023, 28, 881. [Google Scholar] [CrossRef]
- Corrie, L.; Gulati, M.; Vishwas, S.; Kapoor, B.; Singh, S.K.; Awasthi, A.; Khursheed, R. Combination therapy of curcumin and fecal microbiota transplant: Potential treatment of polycystic ovarian syndrome. Med. Hypotheses 2021, 154, 110644. [Google Scholar] [CrossRef]
- Corrie, L.; Gulati, M.; Awasthi, A.; Vishwas, S.; Kaur, J.; Khursheed, R.; Kumar, R.; Kumar, A.; Imran, M.; Chellappan, D.K.; et al. Polysaccharide, fecal microbiota, and curcumin-based novel oral colon-targeted solid self-nanoemulsifying delivery system: Formulation, characterization, and in-vitro anticancer evaluation. Mater. Today Chem. 2022, 26, 101165. [Google Scholar] [CrossRef]
- Corrie, L.; Kaur, J.; Awasthi, A.; Vishwas, S.; Gulati, M.; Saini, S.; Kumar, B.; Pandey, N.K.; Gupta, G.; Dureja, H.; et al. Multivariate Data Analysis and Central Composite Design-Oriented Optimization of Solid Carriers for Formulation of Curcumin-Loaded Solid SNEDDS: Dissolution and Bioavailability Assessment. Pharmaceutics 2022, 14, 2395. [Google Scholar] [CrossRef]
- Lei, F.; Li, P.; Chen, T.; Wang, Q.; Wang, C.; Liu, Y.; Deng, Y.; Zhang, Z.; Xu, M.; Tian, J.; et al. Recent advances in curcumin-loaded biomimetic nanomedicines for targeted therapies. J. Drug Deliv. Sci. Technol. 2023, 80, 104200. [Google Scholar] [CrossRef]
- Corrie, L.; Gulati, M.; Awasthi, A.; Vishwas, S.; Kaur, J.; Khursheed, R.; Porwal, O.; Alam, A.; Parveen, S.R.; Singh, H.; et al. Harnessing the dual role of polysaccharides in treating gastrointestinal diseases: As therapeutics and polymers for drug delivery. Chem. Biol. Interact. 2022, 368, 110238. [Google Scholar] [CrossRef]
- Kurczewska, J. Recent Reports on Polysaccharide-Based Materials for Drug Delivery. Polymers 2022, 14, 4189. [Google Scholar] [CrossRef]
- Qiu, A.; Wang, Y.; Zhang, G.; Wang, H. Natural Polysaccharide-Based Nanodrug Delivery Systems for Treatment of Diabetes. Polymers 2022, 14, 3217. [Google Scholar] [CrossRef]
- Chen, L.; He, C.; Zhou, M.; Long, J.; Li, L. Research Progress on the Mechanisms of Polysaccharides against Gastric Cancer. Molecules 2022, 27, 5828. [Google Scholar] [CrossRef]
- Yang, J.; Shen, M.; Wen, H.; Luo, Y.; Huang, R.; Rong, L.; Xie, J. Recent advance in delivery system and tissue engineering applications of chondroitin sulfate. Carbohydr. Polym. 2020, 230, 115650. [Google Scholar] [CrossRef]
- Żak, A.; Łazarski, G.; Wytrwal-Sarna, M.; Jamróz, D.; Górniewicz, M.; Foryś, A.; Trzebicka, B.; Kepczynski, M. Molecular insights into the self-assembly of hydrophobically modified chondroitin sulfate in aqueous media. Carbohydr. Polym. 2022, 297, 119999. [Google Scholar] [CrossRef]
- Shi, X.; Yang, X.; Liu, M.; Wang, R.; Qiu, N.; Liu, Y.; Yang, H.; Ji, J.; Zhai, G. Chondroitin sulfate-based nanoparticles for enhanced chemo-photodynamic therapy overcoming multidrug resistance and lung metastasis of breast cancer. Carbohydr. Polym. 2021, 254, 117459. [Google Scholar] [CrossRef]
- Zhao, L.; Liu, M.; Wang, J.; Zhai, G. Chondroitin sulfate-based nanocarriers for drug/gene delivery. Carbohydr. Polym. 2015, 133, 391–399. [Google Scholar] [CrossRef]
- Abdullah, T.A.; Ibrahim, N.J.; Warsi, M.H. Chondroitin sulfate-chitosan nanoparticles for ocular delivery of bromfenac sodium: Improved permeation, retention, and penetration. Int. J. Pharm. Investig. 2016, 6, 96–105. [Google Scholar] [CrossRef]
- Yu, Z.; Yu, D.; Dong, J.; Xia, W. Ultrasound-reinforced encapsulation of proanthocyanidin by chitosan-chondroitin sulfate nanosystem. Food Hydrocoll. 2022, 132, 107872. [Google Scholar] [CrossRef]
- Jardim, K.V.; Joanitti, G.A.; Azevedo, R.B.; Parize, A.L. Physico-chemical characterization and cytotoxicity evaluation of curcumin loaded in chitosan/chondroitin sulfate nanoparticles. Mater. Sci. Eng. C Mater. Biol. Appl. 2015, 56, 294–304. [Google Scholar] [CrossRef]
- Jardim, K.V.; Siqueira, J.L.N.; Bao, S.N.; Sousa, M.H.; Parize, A.L. The role of the lecithin addition in the properties and cytotoxic activity of chitosan and chondroitin sulfate nanoparticles containing curcumin. Carbohydr. Polym. 2020, 227, 115351. [Google Scholar] [CrossRef]
- Hu, C.-S.; Tang, S.-L.; Chiang, C.-H.; Hosseinkhani, H.; Hong, P.-D.; Yeh, M.-K. Characterization and anti-tumor effects of chondroitin sulfate–chitosan nanoparticles delivery system. J. Nanopart. Res. 2014, 16, 2672. [Google Scholar] [CrossRef]
- Suner, S.S.; Sahiner, M.; Ayyala, R.S.; Sahiner, N. Degradable and Non-Degradable Chondroitin Sulfate Particles with the Controlled Antibiotic Release for Bacterial Infections. Pharmaceutics 2022, 14, 1739. [Google Scholar] [CrossRef]
- Hu, C.S.; Chiang, C.H.; Hong, P.D.; Yeh, M.K. Influence of charge on FITC-BSA-loaded chondroitin sulfate-chitosan nanoparticles upon cell uptake in human Caco-2 cell monolayers. Int. J. Nanomed. 2012, 7, 4861–4872. [Google Scholar] [CrossRef]
- Guan, Z.; Feng, Q. Chitosan and Chitooligosaccharide: The Promising Non-Plant-Derived Prebiotics with Multiple Biological Activities. Int. J. Mol. Sci. 2022, 23, 6761. [Google Scholar] [CrossRef]
- Tabassum, N.; Ahmed, S.; Ali, M.A. Chitooligosaccharides and their structural-functional effect on hydrogels: A review. Carbohydr. Polym. 2021, 261, 117882. [Google Scholar] [CrossRef]
- Yadav, N.; Mudgal, D.; Anand, R.; Jindal, S.; Mishra, V. Recent development in nanoencapsulation and delivery of natural bioactives through chitosan scaffolds for various biological applications. Int. J. Biol. Macromol. 2022, 220, 537–572. [Google Scholar] [CrossRef]
- Sargazi, S.; Siddiqui, B.; Qindeel, M.; Rahdar, A.; Bilal, M.; Behzadmehr, R.; Mirinejad, S.; Pandey, S. Chitosan nanocarriers for microRNA delivery and detection: A preliminary review with emphasis on cancer. Carbohydr. Polym. 2022, 290, 119489. [Google Scholar] [CrossRef]
- Shoueir, K.R.; El-Desouky, N.; Rashad, M.M.; Ahmed, M.K.; Janowska, I.; El-Kemary, M. Chitosan based-nanoparticles and nanocapsules: Overview, physicochemical features, applications of a nanofibrous scaffold, and bioprinting. Int. J. Biol. Macromol. 2021, 167, 1176–1197. [Google Scholar] [CrossRef]
- Cao, R.; Li, X.; Zhou, Z.; Zhao, Z. Synthesis and biophysical analysis of Naringin-Chitooligosaccharide complex. Nat. Prod. Res. 2021, 35, 305–311. [Google Scholar] [CrossRef]
- Cao, R.; Zhao, Y.; Zhou, Z.; Zhao, X. Enhancement of the water solubility and antioxidant activity of hesperidin by chitooligosaccharide. J. Sci. Food Agric. 2018, 98, 2422–2427. [Google Scholar] [CrossRef]
- Cao, R.; Ma, Q.; Fu, Y.; Zhou, Z.; Zhao, X. Preparation, Evaluation and Characterization of Rutin-Chitooligosaccharide Complex. Plant Foods Hum. Nutr. 2019, 74, 328–333. [Google Scholar] [CrossRef]
- Bockuviene, A.; Sereikaite, J. Preparation and characterisation of novel water-soluble beta-carotene-chitooligosaccharides complexes. Carbohydr. Polym. 2019, 225, 115226. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis of Association of Official Analytical Chemists International; AOAC International: Rockville, MD, USA, 2016; Volume 20. [Google Scholar]
- Tiamsai, R.; Waiprib, Y.; Runglerdkriangkrai, J.; Somjit, K. Optimal level of added salt to minimize the risk of associated chemical hazards of small-sized anchovy (Stolephorus indicus) stored at ambient and refrigerated temperatures. Agric. Nat. Resour. 2022, 56, 9–22. [Google Scholar]
- Garnjanagoonchorn, W.; Wongekalak, L.; Engkagul, A. Determination of chondroitin sulfate from different sources of cartilage. Chem. Eng. Process. Process Intensif. 2007, 46, 465–471. [Google Scholar] [CrossRef]
- Sundaresan, G.; Abraham, R.J.J.; Appa Rao, V.; Narendra Babu, R.; Govind, V.; Meti, M.F. Established method of chondroitin sulphate extraction from buffalo (Bubalus bubalis) cartilages and its identification by FTIR. J. Food Sci. Technol. 2018, 55, 3439–3445. [Google Scholar] [CrossRef]
- Korkmaz, K.; Tokur, B. Optimization of hydrolysis conditions for the production of protein hydrolysates from fish wastes using response surface methodology. Food Biosci. 2022, 45, 101312. [Google Scholar] [CrossRef]
- Zhao, L.; Du, J.; Duan, Y.; Zang, Y.; Zhang, H.; Yang, C.; Cao, F.; Zhai, G. Curcumin loaded mixed micelles composed of Pluronic P123 and F68: Preparation, optimization and in vitro characterization. Colloids Surf. B Biointerfaces 2012, 97, 101–108. [Google Scholar] [CrossRef]
- Jampafuang, Y.; Tongta, A.; Waiprib, Y. Impact of Crystalline Structural Differences Between alpha- and beta-Chitosan on Their Nanoparticle Formation Via Ionic Gelation and Superoxide Radical Scavenging Activities. Polymers 2019, 11, 2010. [Google Scholar] [CrossRef]
- Fang, M.; Jin, Y.; Bao, W.; Gao, H.; Xu, M.; Wang, D.; Wang, X.; Yao, P.; Liu, L. In vitro characterization and in vivo evaluation of nanostructured lipid curcumin carriers for intragastric administration. Int. J. Nanomed. 2012, 7, 5395–5404. [Google Scholar] [CrossRef] [Green Version]
- Rezaei, A.; Nasirpour, A. Evaluation of Release Kinetics and Mechanisms of Curcumin and Curcumin-β-Cyclodextrin Inclusion Complex Incorporated in Electrospun Almond Gum/PVA Nanofibers in Simulated Saliva and Simulated Gastrointestinal Conditions. BioNanoScience 2019, 9, 438–445. [Google Scholar] [CrossRef]
- Yeh, M.K.; Cheng, K.M.; Hu, C.S.; Huang, Y.C.; Young, J.J. Novel protein-loaded chondroitin sulfate-chitosan nanoparticles: Preparation and characterization. Acta Biomater. 2011, 7, 3804–3812. [Google Scholar] [CrossRef]
- Srihera, N.; Li, Y.; Zhang, T.T.; Wang, Y.M.; Yanagita, T.; Waiprib, Y.; Xue, C.H. Preparation and Characterization of Astaxanthin-loaded Liposomes Stabilized by Sea Cucumber Sulfated Sterols Instead of Cholesterol. J. Oleo Sci. 2022, 71, 401–410. [Google Scholar] [CrossRef]
- Li, W.; Kobayashi, T.; Meng, D.; Miyamoto, N.; Tsutsumi, N.; Ura, K.; Takagi, Y. Free radical scavenging activity of type II collagen peptides and chondroitin sulfate oligosaccharides from by-products of mottled skate processing. Food Biosci. 2021, 41, 100991. [Google Scholar] [CrossRef]
- Herpandi, N.H.; Rosma, A.; Wan Nadiah, W.A. The Tuna Fishing Industry: A New Outlook on Fish Protein Hydrolysates. Compr. Rev. Food Sci. Food Saf. 2011, 10, 195–207. [Google Scholar] [CrossRef]
- Li, W.; Liu, Y.; Jiang, W.; Yan, X. Proximate Composition and Nutritional Profile of Rainbow Trout (Oncorhynchus mykiss) Heads and Skipjack tuna (Katsuwonus pelamis) Heads. Molecules 2019, 24, 3189. [Google Scholar] [CrossRef]
- Oliveira, D.; Bernardi, D.; Drummond, F.; Dieterich, F.; Boscolo, W.; Leivas, C.; Kiatkoski, E.; Waszczynskyj, N. Potential Use of Tuna (Thunnus albacares) by-product: Production of Antioxidant Peptides and Recovery of Unsaturated Fatty Acids from Tuna Head. Int. J. Food Eng. 2017, 13, 20150365. [Google Scholar] [CrossRef]
- Arima, K.; Fujita, H.; Toita, R.; Imazu-Okada, A.; Tsutsumishita-Nakai, N.; Takeda, N.; Nakao, Y.; Wang, H.; Kawano, M.; Matsushita, K.; et al. Amounts and compositional analysis of glycosaminoglycans in the tissue of fish. Carbohydr. Res. 2013, 366, 25–32. [Google Scholar] [CrossRef]
- Maccari, F.; Galeotti, F.; Volpi, N. Isolation and structural characterization of chondroitin sulfate from bony fishes. Carbohydr. Polym. 2015, 129, 143–147. [Google Scholar] [CrossRef]
- Yuvashri, U.; Kanchana, S.; Abirami, A.; Naidu, K.; Rajarao, G.K.; Arumugam, M. Chondroitin Sulfate from Marine Invertebrates. In Encyclopedia of Marine Biotechnology; Wiley Online Library: New York, NY, USA, 2020; pp. 1051–1063. [Google Scholar]
- Thomas, D.L.; Kp, D.M.; Mathew, D.S. Fish gills of Thunnus albacares: A novel source of chondroitin sulphate glycosaminoglycans. Int. J. Fish. Aquat. Stud. 2021, 9, 146–152. [Google Scholar] [CrossRef]
- Krichen, F.; Bougatef, H.; Capitani, F.; Ben Amor, I.; Koubaa, I.; Gargouri, J.; Maccari, F.; Mantovani, V.; Galeotti, F.; Volpi, N.; et al. Purification and structural elucidation of chondroitin sulfate/dermatan sulfate from Atlantic bluefin tuna (Thunnus thynnus) skins and their anticoagulant and ACE inhibitory activities. RSC Adv. 2018, 8, 37965–37975. [Google Scholar] [CrossRef]
- Yang, J.; Shen, M.; Wu, T.; Chen, X.; Wen, H.; Xie, J. Physicochemical, structural characterization, and antioxidant activities of chondroitin sulfate from Oreochromis niloticus bones. Food Sci. Hum. Wellness 2023, 12, 1102–1108. [Google Scholar] [CrossRef]
- Valcarcel, J.; Garcia, M.R.; Sampayo, L.F.; Vazquez, J.A. Marine chondroitin sulfate of defined molecular weight by enzymatic depolymerization. Carbohydr. Polym. 2020, 229, 115450. [Google Scholar] [CrossRef]
- Zhou, C.; Mi, S.; Li, J.; Gao, J.; Wang, X.; Sang, Y. Purification, characterisation and antioxidant activities of chondroitin sulphate extracted from Raja porosa cartilage. Carbohydr. Polym. 2020, 241, 116306. [Google Scholar] [CrossRef]
- Dong, F.K.; Quan, X.G.; Wang, Q.B.; Liu, Z.M.; Cui, T.; Wang, W.J.; Tang, D.M.; Zhang, R.M.; Zhang, C.; Wang, H.Y.; et al. Purification, structural characterization, and anticoagulant activity evaluation of chondroitin sulfate from codfish (Gadus macrocephalus) bones. Int. J. Biol. Macromol. 2022, 210, 759–767. [Google Scholar] [CrossRef]
- Wang, K.; Qi, L.; Zhao, L.; Liu, J.; Guo, Y.; Zhang, C. Degradation of chondroitin sulfate: Mechanism of degradation, influence factors, structure-bioactivity relationship and application. Carbohydr. Polym. 2023, 301, 120361. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhang, S.; Ma, M.; Xu, Y.; Wang, D. Delivery of curcumin by shellac encapsulation: Stability, bioaccessibility, freeze-dried redispersibility, and solubilization. Food Chem. X 2022, 15, 100431. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, H.; Xu, C.; Gu, L. A review: Using nanoparticles to enhance absorption and bioavailability of phenolic phytochemicals. Food Hydrocoll. 2015, 43, 153–164. [Google Scholar] [CrossRef]
- Lee, J.S.; Hong, D.Y.; Kim, E.S.; Lee, H.G. Improving the water solubility and antimicrobial activity of silymarin by nanoencapsulation. Colloids Surf. B Biointerfaces 2017, 154, 171–177. [Google Scholar] [CrossRef]
- Del Prado-Audelo, M.L.; Magaña, J.J.; Mejía-Contreras, B.A.; Borbolla-Jiménez, F.V.; Giraldo-Gomez, D.M.; Piña-Barba, M.C.; Quintanar-Guerrero, D.; Leyva-Gómez, G. In vitro cell uptake evaluation of curcumin-loaded PCL/F68 nanoparticles for potential application in neuronal diseases. J. Drug Deliv. Sci. Technol. 2019, 52, 905–914. [Google Scholar] [CrossRef]
- Sarabandi, K.; Gharehbeglou, P.; Jafari, S.M. Scanning electron microscopy (SEM) of nanoencapsulated food ingredients. In Characterization of Nanoencapsulated Food Ingredients; Academic Press: Cambridge, MA, USA, 2020; pp. 83–130. [Google Scholar]
- Feyzioglu, G.C.; Tornuk, F. Development of chitosan nanoparticles loaded with summer savory (Satureja hortensis L.) essential oil for antimicrobial and antioxidant delivery applications. LWT 2016, 70, 104–110. [Google Scholar] [CrossRef]
- Wang, J.; Wang, L.; Li, Y.; Wang, X.; Tu, P. Apically targeted oral micelles exhibit highly efficient intestinal uptake and oral absorption. Int. J. Nanomed. 2018, 13, 7997–8012. [Google Scholar] [CrossRef]
- Wang, J.; Ma, W.; Tu, P. The mechanism of self-assembled mixed micelles in improving curcumin oral absorption: In vitro and in vivo. Colloids Surf. B Biointerfaces 2015, 133, 108–119. [Google Scholar] [CrossRef]
- Malekjani, N.; Jafari, S.M. Release modeling of nanoencapsulated food ingredients by empirical and semiempirical models. In Release and Bioavailability of Nanoencapsulated Food Ingredients; Academic Press: Cambridge, MA, USA, 2020; pp. 211–246. [Google Scholar]
- Zhu, W.; Ji, Y.; Wang, Y.; He, D.; Yan, Y.; Su, N.; Zhang, C.; Xing, X.H. Structural characterization and in vitro antioxidant activities of chondroitin sulfate purified from Andrias davidianus cartilage. Carbohydr. Polym. 2018, 196, 398–404. [Google Scholar] [CrossRef]
- Jafari, H.; Bernaerts, K.V.; Dodi, G.; Shavandi, A. Chitooligosaccharides for wound healing biomaterials engineering. Mater. Sci. Eng. C Mater. Biol. Appl. 2020, 117, 111266. [Google Scholar] [CrossRef]
- Lin, D.; Xiao, L.; Qin, W.; Loy, D.A.; Wu, Z.; Chen, H.; Zhang, Q. Preparation, characterization and antioxidant properties of curcumin encapsulated chitosan/lignosulfonate micelles. Carbohydr. Polym. 2022, 281, 119080. [Google Scholar] [CrossRef]
- Valencia, M.S.; Silva Júnior, M.F.d.; Xavier-Júnior, F.H.; Veras, B.d.O.; Albuquerque, P.B.S.d.; Borba, E.F.d.O.; Silva, T.G.d.; Xavier, V.L.; Souza, M.P.d.; Carneiro-da-Cunha, M.d.G. Characterization of curcumin-loaded lecithin-chitosan bioactive nanoparticles. Carbohydr. Polym. Technol. Appl. 2021, 2, 100119. [Google Scholar] [CrossRef]
- Wang, H.; Betti, M. Sulfated glycosaminoglycan-derived oligosaccharides produced from chicken connective tissue promote iron uptake in a human intestinal Caco-2 cell line. Food Chem. 2017, 220, 460–469. [Google Scholar] [CrossRef]
- Tan, C.; Xie, J.; Zhang, X.; Cai, J.; Xia, S. Polysaccharide-based nanoparticles by chitosan and gum arabic polyelectrolyte complexation as carriers for curcumin. Food Hydrocoll. 2016, 57, 236–245. [Google Scholar] [CrossRef]








| Parameters | Skipjack Tuna Heads | Yellowfin Tuna Heads | ||
|---|---|---|---|---|
| In the Present Study | Li et al. [71] | In the Present Study | Oliveira et al. [72] | |
| Moisture | 66.70 ± 0.70 | 75.6 ± 0.5 | 65.09 ± 1.31 | 70.1 |
| Protein | 18.35 ± 0.23 a | 18 ± 3 | 16.96 ± 0.33 b | 15.1 |
| Lipid | 3.64 ± 0.21 | 4.8 ± 0.5 | 4.12 ± 0.63 | 7.1 |
| Ash | 9.71 ± 0.22 a | 3.88 ± 0.08 | 8.05 ± 0.31 b | 5.18 |
| Zero Model | Higuchi Model | Kopcha Model | Korsmeyer–Peppas Model | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R2 | R2 | R2 | R2 | |||||||
| CU | 4.5645 | 0.4498 | 453.7787 | 0.7076 | 21.3021 | 0.0001 | 0.7076 | 3.17×107 | 0.2171 | 0.8808 |
| CHCU | 1.9139 | 0.8707 | 66.9977 | 0.9658 | 8.1239 | 0.0002 | 0.9656 | 116.35 | 0.4610 | 0.9671 |
| CHCSCU | 2.1641 | 0.8486 | 84.9332 | 0.9471 | 9.2159 | 0.0011 | 0.9471 | 193.36 | 0.4471 | 0.9496 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waiprib, Y.; Ingrungruengluet, P.; Worawattanamateekul, W. Nanoparticles Based on Chondroitin Sulfate from Tuna Heads and Chitooligosaccharides for Enhanced Water Solubility and Sustained Release of Curcumin. Polymers 2023, 15, 834. https://doi.org/10.3390/polym15040834
Waiprib Y, Ingrungruengluet P, Worawattanamateekul W. Nanoparticles Based on Chondroitin Sulfate from Tuna Heads and Chitooligosaccharides for Enhanced Water Solubility and Sustained Release of Curcumin. Polymers. 2023; 15(4):834. https://doi.org/10.3390/polym15040834
Chicago/Turabian StyleWaiprib, Yaowapha, Pattarachat Ingrungruengluet, and Wanchai Worawattanamateekul. 2023. "Nanoparticles Based on Chondroitin Sulfate from Tuna Heads and Chitooligosaccharides for Enhanced Water Solubility and Sustained Release of Curcumin" Polymers 15, no. 4: 834. https://doi.org/10.3390/polym15040834