Insights into Fluctuations of Structure of Proteins: Significance of Intermediary States in Regulating Biological Functions
Abstract
:1. Introduction
2. Intermediate States of Proteins and Their Types
2.1. Intermediate States Characterized under In Vitro Conditions
| S. No. | Protein | State Type | Conditions | Techniques Exploited | Ref. |
|---|---|---|---|---|---|
| 1. | Apo-α-lactalbumin | MG | At neutral pH (7.6) and low ionic strength | Scanning microcalorimeter | [42] |
| 2. | Apo-α-lactalbumin | MG | The transition around 25–30 °C at pH 8.1 in the presence of 10 mM borate and 1 mM EGTA | Intrinsic protein fluorescence, circular dichroism (CD), and differential scanning microcalorimetry (DSC) | [7] |
| 3. | α-Lactalbumin | MG | Guanidinium chloride (GdmCl)-induced (1.8 M) and 1 mM Ca2+ at 4.5 °C, pH 7.0 in the presence of 0.05 M sodium chloride (NaCl) and 0.05 M sodium cacodylate | Circular dichroism (CD) spectroscopy and nuclear magnetic resonance | [81] |
| 4. | Myoglobin | MG | PEG 10 (300 mg mL−1) at pH 7.0 and 25 °C | Absorption, fluorescence and CD spectroscopy, ANS binding, dynamic light scattering (DLS), FTIR, isothermal titration calorimetry (ITC) | [30] |
| 5. | Myoglobin | MG | Ficoll 70 (300 mg mL−1) at pH 7.0 and 25 °C | CD spectroscopy, intrinsic and ANS fluorescence, DLS, and ITC measurements | [29] |
| 6. | Myoglobin | PMG | PEG 400 (320 mg mL−1) at pH 7.0 and 25 °C | CD spectroscopy, intrinsic and ANS fluorescence, DLS, and ITC measurements | [33] |
| 6. | Myoglobin | MG | Around 300 K (26.85°C) −500 K (226.85 °C), apo-Mb like intermediate state for 2–9 ns (nanoseconds) at pH 7.0 | In silico method (i.e., molecular dynamic (MD) simulations) | [82] |
| 5. | Myoglobin | MG | Cobalt(III) induced (10 μM) in 0.01 M sodium phosphate buffer solution at pH 6.5 and 25 °C | UV–VIS absorption and CD spectroscopy | [83] |
| 7. | Myoglobin | MG | 4% (/v) HFIP (aqueous hexafluoroisopropanol) at pH 4.0 | CD spectroscopy | [84] |
| 9. | Apo-myoglobin (Apo-Mb) | MG | Site mutagenesis studies at pH 7.0 and pH 3.0 | Fluorescence and CD spectroscopy | [85] |
| 10. | Apo-myoglobin (mutants) | MG | Mutation in apo-Mb (S108L, F123W, F123G, and A130S) in the presence of 10 mM sodium acetate buffer at 0 °C around acidic pH | Circular dichroism (CD) spectroscopy, nuclear magnetic resonance | [26] |
| 11. | Apo-myoglobin | MG | Acid-induced unfolding at 0 °C, 2 mM sodium citrate in the presence of various urea concentrations | Circular dichroism (CD) spectroscopy, nuclear magnetic resonance | [86] |
| 12. | Apo-myoglobin | PMG | In the presence of different anions (100 mM trifluoroacetate) at pH 2.0 and 25 °C | Tryptophan and ANS binding fluorescence, CD spectroscopy, FTIR, small-angle X-ray scattering, and DLS | [87] |
| 16. | Cytochrome c | MG | PEG 400 induced at pH 7.0 and 25 °C | Absorption, fluorescence and CD spectroscopy, DLS, and ITC measurements | [88] |
| 17. | Cytochrome c | MG | Induced by LiClO4 (1.85–3.3 M) at pH 6.0 and 25 °C | CD spectroscopy, intrinsic and ANS fluorescence, and DLS and intrinsic viscosity measurements | [32] |
| 18. | Yeast iso-1-cytochrome c and its deletants | PMG | Induced by LiCl at pH 6.5 at 25 °C | Absorption, fluorescence, and CD spectroscopy and DLS measurements | [55] |
| 19. | Cytochrome c (mutant Leu94Gly) | PMG | Induced by LiCl at pH 6.5 at 25 °C | Tryptophan fluorescence, ANS binding, CD spectroscopy, and DLS measurements | [57] |
| 20. | Cytochrome c | PMG | NaCl-induced L94G mutation at pH 2 and 25 °C | CD spectroscopy, intrinsic and ANS fluorescence, and DLS measurements | [19] |
| 13. | Cytochrome c | MG | Mutation of Leu94Gly at pH 6.0 and 25 °C | CD spectroscopy, intrinsic and ANS fluorescence, and DLS measurements | [19] |
| 14. | Cytochrome c | MG | Leu94 by Val and Ile, at pH 6.0 and 25 °C | Intrinsic fluorescence and CD spectroscopy and differential scanning microcalorimetry (DSC) | [22] |
| 15. | Cytochrome c | MG | Leu94 by Phe at pH 6.0 and 25 °C | Intrinsic fluorescence and CD spectroscopy, ANS binding, and DSC measurements | [89] |
| 20. | Cytochrome c | PMG | NaCl-induced L94G mutation at pH 2 and 25 °C | CD spectroscopy, intrinsic and ANS fluorescence, and DLS measurements | [19] |
| 21. | Cytochrome c | MG | Polyol-induced (ethylene glycol, glycerol, erythritol, xylitol, sorbitol, and inositol) at pH 2.0 | Circular dichroism (CD) spectroscopy, partial specific volume, adiabatic compressibility, and DSC | [90] |
| 22. | Yeast iso-1-cytochrome c and its deletants | MG | In the presence of 0.33 M Na2SO4 at pH 2.1 | Absorption, fluorescence, and CD spectroscopy and DLS measurements | [55] |
| 23. | Cytochrome c | MG | Sodium perchlorate stabilized at pH 1.8 | Isothermal titration calorimetry, CD spectroscopy and DSC | [91] |
| 24. | Sheep serum albumin | MG | GdmCl (2.38 M)-induced denaturation and urea (4.2–4.7 M)-induced denaturationin10 mM Tris-HCl buffer at pH 7.4 and 25 °C | Intrinsic and ANS binding fluorescence, CD spectroscopy, and DLS measurements | [92] |
| 25. | Bovine carbonic anhydrase B | PMG | At 4 °C in 0.1 M sodium phosphate buffer (pH 6.8) in the presence of GdmCl concentrations | Tryptophan and ANS binding fluorescence, CD spectroscopy, size-exclusion chromatography (SEC-FPLC) | [28] |
| 26. | GlutaminyltRNA synthetase (GlnRS) | PMG | Induced by 0.25 M potassium L-glutamate (natural osmolyte) in the presence of urea, 0.1 M Tris-HCl buffer of pH 7.5 at 25 °C | Tryptophan and ANS binding fluorescence, CD spectroscopy, and DLS measurements | [93] |
| 27. | Recombinant human Stefan B | MG-states (G, A, and T) | G-state:in the presence of 1.7 M GdmCl (pH 8, 25 °C), A-state: at pH 4 (0.6 M GdmHCl, 25 °C), and T-state: formed above 68 °C | UV–VIS absorption and CD spectroscopy | [94] |
| 28. | Pancreatic trypsin inhibitor (BPTI) | MG | Five MD simulations (lasting up to 550 ps) were performed: native BPTI at 298 K (25 °C) and 423 K (150 °C); reduced BPTI at 298 K (25 °C), 423 K (150 °C), and 498 K (225 °C); all simulations were carried out in a bath of water molecules with mobile counter ions | MD simulations | [95] |
| 29. | Casein | PMG and MG | Physiological conditions (around pH 7) | Raman spectroscopy, FTIR, DLS measurements, and molecular kinetics | [96] |
| 30. | Lysozyme | MG | At pH 2.0 | Hydrogen exchange measurements, NMR, molecular graphics by MolScript | [97] |
| 31. | Ribonuclease A | MG | At low pH (1.5—3.8) and 65 °C | Quenched flow methods, CD spectroscopy, pulsed H/D-exchange, and 2 D 1H NMR spectroscopy | [98] |
| 32. | Ubiquitin | MG | At pH 2.0 and 25 °C in the presence of 60% methanol and 40% water | Pulsed H/D-exchange, NMR | [99] |
| 33. | Zinc finger protein Ros87 | Metal-binding intermediate | At pH 6.5 and temperature range of 25—99 °C (observed at 70 °C by NMR) | CD, DSC, NMR | [21] |
| 34. | Apoflavodoxin | Thermal intermediate | At pH 7.0 and 95 °C | Atomistic multi-microsecond-scale molecular dynamics (MD) simulations, small-angle X-ray scattering, near-UV absorbance spectra | [100] |
| 35. | Bovine serum albumin (BSA) | MG | In the presence of ANS and pyrene at pH 4.2 | ANS fluorescence (supplemented by CD spectroscopy, light scattering, and analytical centrifugation) | [101] |
| 36. | Staphylococcal nuclease (SNase) | Three different partially folded intermediates (A states: A1, A2, and A3) | Induced by anions: A states are stabilized by: (1) A1: induced by chloride (600 mM) or sulfate (100 mM): 50% native-like structure (2) A2: Induced by trifluoroacetate (300 mM): 70% native-like structure (3) A3: trichloroacetate (50 mM): fully native-like structure | CD and small-angle X-ray scattering (SAXS) | [102] |
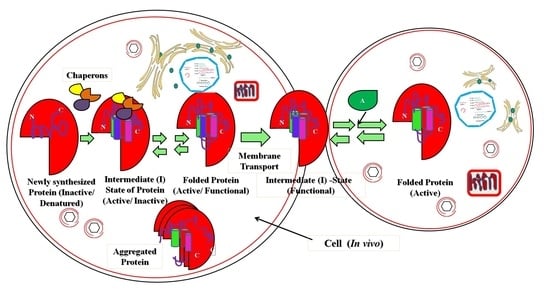
2.2. Significance of Intermediary States under In Vivo Conditions
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Declarations
References
- Parray, Z.A.; Ahmad, F.; Alajmi, M.F.; Hussain, A.; Hassan, M.I.; Islam, A. Interaction of polyethylene glycol with cytochrome c investigated via in vitro and in silico approaches. Sci. Rep. 2021, 11, 6475. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Johnson, A.; Lewis, J. The Transport of Proteins into Mitochondria and Chloroplasts. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. In vivo aspects of protein folding and quality control. Science 2016, 353, aac4354. [Google Scholar] [CrossRef] [PubMed]
- Banks, A.; Qin, S.; Weiss, K.L.; Stanley, C.B.; Zhou, H.X. Intrinsically Disordered Protein Exhibits Both Compaction and Expansion under Macromolecular Crowding. Biophys. J. 2018, 114, 1067–1079. [Google Scholar] [CrossRef] [PubMed]
- Tsytlonok, M.; Itzhaki, L.S. The how’s and why’s of protein folding intermediates. Arch. Biochem. Biophys. 2013, 531, 14–23. [Google Scholar] [CrossRef]
- Dyson, H.J.; Wright, P.E. Unfolded proteins and protein folding studied by NMR. Chem. Rev. 2004, 104, 3607–3622. [Google Scholar] [CrossRef]
- Veprintsev, D.B.; Permyakov, S.E.; Permyakov, E.A.; Rogov, V.V.; Cawthern, K.M.; Berliner, L.J. Cooperative thermal transitions of bovine and human apo-alpha-lactalbumins: Evidence for a new intermediate state. FEBS Lett. 1997, 412, 625–628. [Google Scholar] [CrossRef] [Green Version]
- Santucci, R.; Polticelli, F.; Sinibaldi, F.; Fiorucci, L. Chapter 14—Role of Intermediate States in Protein Folding and Misfolding. In Recent Advances in Medicinal Chemistry; Atta ur, R., Choudhary, M.I., Perry, G., Eds.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 433–455. [Google Scholar]
- Boesch, C.; Bundi, A.; Oppliger, M.; Wuthrich, K. 1H nuclear-magnetic-resonance studies of the molecular conformation of monomeric glucagon in aqueous solution. Eur. J. Biochem. 1978, 91, 209–214. [Google Scholar] [CrossRef]
- Daily, S. How a Membrane Protein Can Move Both Lipids and Ions. Science Daily, University of Groningen. 2019. Available online: https://www.sciencedaily.com/releases/2019/03/190312075928.htm (accessed on 20 February 2022).
- Alvadia, C.; Lim, N.K.; Clerico Mosina, V.; Oostergetel, G.T.; Dutzler, R.; Paulino, C. Cryo-EM structures and functional characterization of the murine lipid scramblase TMEM16F. Elife 2019, 8, e44365. [Google Scholar] [CrossRef]
- Hou, X.; Aguilar, M.I.; Small, D.H. Transthyretin and familial amyloidotic polyneuropathy. Recent progress in understanding the molecular mechanism of neurodegeneration. FEBS J. 2007, 274, 1637–1650. [Google Scholar] [CrossRef] [Green Version]
- Bellotti, V.; Mangione, P.; Merlini, G. Review: Immunoglobulin light chain amyloidosis--the archetype of structural and pathogenic variability. J. Struct. Biol. 2000, 130, 280–289. [Google Scholar] [CrossRef]
- Eakin, C.; Miranker, A. From chance to frequent encounters: Origins of β2-microglobulin fibrillogenesis. Biochim. Biophys. Acta 2005, 1753, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.W. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr. Opin. Struct. Biol. 1998, 8, 101–106. [Google Scholar] [CrossRef]
- Fandrich, M. On the structural definition of amyloid fibrils and other polypeptide aggregates. Cell. Mol. Life Sci. 2007, 64, 2066–2078. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ding, F.; Nie, H.; Serohijos, A.W.; Sharma, S.; Wilcox, K.C.; Yin, S.; Dokholyan, N.V. Protein folding: Then and now. Arch. Biochem. Biophys. 2008, 469, 4–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eaton, W.A.; Thompson, P.A.; Chan, C.-K.; Hage, S.J.; Hofrichter, J. Fast events in protein folding. Structure 1996, 4, 1133–1139. [Google Scholar] [CrossRef] [Green Version]
- Alam Khan, M.K.; Das, U.; Rahaman, M.H.; Hassan, M.I.; Srinivasan, A.; Singh, T.P.; Ahmad, F. A single mutation induces molten globule formation and a drastic destabilization of wild-type cytochrome c at pH 6.0. J. Biol. Inorg. Chem. 2009, 14, 751–760. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Varadarajan, R. Packing in molten globules and native states. Curr. Opin. Struct. Biol. 2013, 23, 11–21. [Google Scholar] [CrossRef]
- Grazioso, R.; García-Viñuales, S.; D’Abrosca, G.; Baglivo, I.; Pedone, P.V.; Milardi, D.; Fattorusso, R.; Isernia, C.; Russo, L.; Malgieri, G. The change of conditions does not affect Ros87 downhill folding mechanism. Sci. Rep. 2020, 10, 21067. [Google Scholar] [CrossRef]
- Khan, S.H.; Islam, A.; Hassan, M.I.; Sharma, S.; Singh, T.P.; Ahmad, F. Effect of conservative mutations (L94V and L94I) on the structure and stability of horse cytochrome c. Arch. Biochem. Biophys. 2017, 633, 40–49. [Google Scholar] [CrossRef]
- Sharma, R.; Kishore, N. Thermodynamic insights into the binding of ANS with the salt induced molten globule states of cytochrome c. J. Chem. Thermodyn. 2009, 41, 342–348. [Google Scholar] [CrossRef]
- Naeem, A.; Khan, R.H. Characterization of molten globule state of cytochrome c at alkaline, native and acidic pH induced by butanol and SDS. Int. J. Biochem. Cell Biol. 2004, 36, 2281–2292. [Google Scholar] [CrossRef] [PubMed]
- Grandori, R. Detecting equilibrium cytochrome c folding intermediates by electrospray ionisation mass spectrometry: Two partially folded forms populate the molten-globule state. Protein Sci. 2002, 11, 453–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughson, F.M.; Wright, P.E.; Baldwin, R.L. Structural characterization of a partly folded apomyoglobin intermediate. Science 1990, 249, 1544–1548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Semisotnov, G.V.; Rodionova, N.A.; Razgulyaev, O.I.; Uversky, V.N.; Gripas, A.F.; Gilmanshin, R.I. Study of the “molten globule” intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers 1991, 31, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Ptitsyn, O.B. Further evidence on the equilibrium “pre-molten globule state”: Four-state guanidinium chloride-induced unfolding of carbonic anhydrase B at low temperature. J. Mol. Biol. 1996, 255, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Nasreen, K.; Ahamad, S.; Ahmad, F.; Hassan, M.I.; Islam, A. Macromolecular crowding induces molten globule state in the native myoglobin at physiological pH. Int. J. Biol. Macromol. 2018, 106, 130–139. [Google Scholar] [CrossRef]
- Parray, Z.A.; Shahid, S.; Ahmad, F.; Hassan, M.I.; Islam, A. Characterization of intermediate state of myoglobin in the presence of PEG 10 under physiological conditions. Int. J. Biol. Macromol. 2017, 99, 241–248. [Google Scholar] [CrossRef]
- Khan, M.K.; Rahaman, H.; Ahmad, F. Conformation and thermodynamic stability of pre-molten and molten globule states of mammalian cytochromes-c. Metallomics 2011, 3, 327–338. [Google Scholar] [CrossRef]
- Moza, B.; Qureshi, S.H.; Islam, A.; Singh, R.; Anjum, F.; Moosavi-Movahedi, A.A.; Ahmad, F. A unique molten globule state occurs during unfolding of cytochrome c by LiClO4 near physiological pH and temperature: Structural and thermodynamic characterization. Biochemistry 2006, 45, 4695–4702. [Google Scholar] [CrossRef]
- Parray, Z.A.; Ahamad, S.; Ahmad, F.; Hassan, M.I.; Islam, A. First evidence of formation of pre-molten globule state in myoglobin: A macromolecular crowding approach towards protein folding in vivo. Int. J. Biol. Macromol. 2019, 126, 1288–1294. [Google Scholar] [CrossRef]
- Parray, Z.A.; Hassan, M.I.; Ahmad, F.; Islam, A. Amphiphilic nature of polyethylene glycols and their role in medical research. Polym. Test. 2020, 82, 106316. [Google Scholar] [CrossRef]
- Naiyer, A.; Hassan, M.I.; Islam, A.; Sundd, M.; Ahmad, F. Structural characterization of MG and pre-MG states of proteins by MD simulations, NMR, and other techniques. J. Biomol. Struct. Dyn. 2015, 33, 2267–2284. [Google Scholar] [CrossRef] [PubMed]
- Arai, M.; Kuwajima, K. Role of the molten globule state in protein folding. Adv. Protein Chem. 2000, 53, 209–282. [Google Scholar]
- Kim, P.S.; Baldwin, R.L. Intermediates in the folding reactions of small proteins. Annu. Rev. Biochem. 1990, 59, 631–660. [Google Scholar] [CrossRef]
- Baldwin, R.L. Experimental studies of pathways of protein folding. Ciba Found. Symp. 1991, 161, 190–201. [Google Scholar]
- Ptitsyn, O.B.; Uversky, V.N. The molten globule is a third thermodynamical state of protein molecules. FEBS Lett. 1994, 341, 15–18. [Google Scholar] [CrossRef] [Green Version]
- Kuwajima, K.; Nitta, K.; Yoneyama, M.; Sugai, S. Three-state denaturation of alpha-lactalbumin by guanidine hydrochloride. J. Mol. Biol. 1976, 106, 359–373. [Google Scholar] [CrossRef]
- Ohgushi, M.; Wada, A. ‘Molten-globule state’: A compact form of globular proteins with mobile side-chains. FEBS Lett. 1983, 164, 21–24. [Google Scholar] [CrossRef] [Green Version]
- Yutani, K.; Ogasahara, K.; Kuwajima, K. Absence of the thermal transition in apo-α-lactalbumin in the molten globule state: A study by differential scanning microcalorimetry. J. Mol. Biol. 1992, 228, 347–350. [Google Scholar] [CrossRef]
- Baldwin, R.L.; Rose, G.D. Molten globules, entropy-driven conformational change and protein folding. Curr. Opin. Struct. Biol. 2013, 23, 4–10. [Google Scholar] [CrossRef]
- Mezei, M. Discriminatory power of stoichiometry-driven protein folding? J. Biomol. Struct. Dyn. 2011, 28, 625–626. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, R. Is protein folding still a challenge? J. Biomol. Struct. Dyn. 2011, 28, 639–640. [Google Scholar] [CrossRef] [PubMed]
- Kuwajima, K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins 1989, 6, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Baum, J.; Dobson, C.M.; Evans, P.A.; Hanley, C. Characterization of a partly folded protein by NMR methods: Studies on the molten globule state of guinea pig alpha-lactalbumin. Biochemistry 1989, 28, 7–13. [Google Scholar] [CrossRef]
- Jeng, M.F.; Englander, S.W.; Elove, G.A.; Roder, H.; Wand, A.J. Structural description of acid-denatured cytochrome c by hydrogen exchange and 2D NMR. Biochemistry 1990, 29, 10433–10437. [Google Scholar] [CrossRef]
- Hamada, D.; Kuroda, Y.; Kataoka, M.; Aimoto, S.; Yoshimura, T.; Goto, Y. Role of heme axial ligands in the conformational stability of the native and molten globule states of horse cytochrome c. J. Mol. Biol. 1996, 256, 172–186. [Google Scholar] [CrossRef]
- Uversky, V.N.; Ptitsyn, O.B. “Partly Folded” State, a New Equilibrium State of Protein Molecules: Four-State Guanidinium Chloride-Induced Unfolding of. beta.-Lactamase at Low Temperature. Biochemistry 1994, 33, 2782–2791. [Google Scholar] [CrossRef]
- Uversky, V.N.; Li, J.; Fink, A.L. Metal-triggered structural transformations, aggregation, and fibrillation of human alpha-synuclein. A possible molecular NK between Parkinson’s disease and heavy metal exposure. J. Biol. Chem. 2001, 276, 44284–44296. [Google Scholar] [CrossRef] [Green Version]
- Ptitsyn, O.B. Protein evolution and protein folding: Non-functional conserved residues and their probable role. In Proceedings of the Pacific Symposium on Biocomputing, PSB 1999, The Big Island, HI, USA, 4–9 January 1999; pp. 494–504. [Google Scholar] [CrossRef] [Green Version]
- Sackett, D.L.; Wolff, J. Nile red as a polarity-sensitive fluorescent probe of hydrophobic protein surfaces. Anal. Biochem. 1987, 167, 228–234. [Google Scholar] [CrossRef]
- Jeng, M.F.; Englander, S.W. Stable submolecular folding units in a non-compact form of cytochrome c. J. Mol. Biol. 1991, 221, 1045–1061. [Google Scholar] [CrossRef]
- Haque, M.A.; Ubaid-Ullah, S.; Zaidi, S.; Hassan, M.I.; Islam, A.; Batra, J.K.; Ahmad, F. Characterization of pre-molten globule state of yeast iso-1-cytochrome c and its deletants at pH 6.0 and 25 °C. Int. J. Biol. Macromol. 2015, 72, 1406–1418. [Google Scholar] [CrossRef] [PubMed]
- Tcherkasskaya, O.; Uversky, V.N. Polymeric aspects of protein folding: A brief overview. Protein Pept. Lett. 2003, 10, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Alam Khan, M.K.; Rahaman, M.H.; Hassan, M.I.; Singh, T.P.; Moosavi-Movahedi, A.A.; Ahmad, F. Conformational and thermodynamic characterization of the premolten globule state occurring during unfolding of the molten globule state of cytochrome c. J. Biol. Inorg. Chem. 2010, 15, 1319–1329. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Li, J.; Fink, A.L. Evidence for a partially folded intermediate in alpha-synuclein fibril formation. J. Biol. Chem. 2001, 276, 10737–10744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumari, N.; Dubey, V.; Jagannadham, M. SDS Induced Refolding of Pre-molten Globule State of Cryptolepain: Differences in Chemical and Temperature-Induced Equilibrium Unfolding of the Protein in SDS-Induced State. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2013, 83, 71–80. [Google Scholar] [CrossRef]
- Uversky, V.N. Size-exclusion chromatography in structural analysis of intrinsically disordered proteins. Methods Mol. Biol. 2012, 896, 179–194. [Google Scholar]
- Uversky, V.N.; Fink, A.L. Do protein molecules have a native-like topology in the pre-molten globule state? Biochemistry 1999, 64, 552–555. [Google Scholar]
- Uversky, V.N. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002, 11, 739–756. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N. What does it mean to be natively unfolded? Eur. J. Biochem. 2002, 269, 2–12. [Google Scholar] [CrossRef]
- Uversky, V.N.; Narizhneva, N.V.; Kirschstein, S.O.; Winter, S.; Lober, G. Conformational transitions provoked by organic solvents in beta-lactoglobulin: Can a molten globule like intermediate be induced by the decrease in dielectric constant? Fold. Des. 1997, 2, 163–172. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N. Protein folding revisited. A polypeptide chain at the folding-misfolding-nonfolding cross-roads: Which way to go? Cell. Mol. Life Sci. 2003, 60, 1852–1871. [Google Scholar] [CrossRef] [PubMed]
- Parray, Z.A.; Naqvi, A.A.T.; Ahmad, F.; Hassan, M.I.; Islam, A. Characterization of different intermediate states in myoglobin induced by polyethylene glycol: A process of spontaneous molecular self-organization foresees the energy landscape theory via in vitro and in silico approaches. J. Mol. Liq. 2021, 342, 117502. [Google Scholar] [CrossRef]
- Ghosh, D.K.; Ranjan, A. The metastable states of proteins. Protein Sci. 2020, 29, 1559–1568. [Google Scholar] [CrossRef] [PubMed]
- Parak, F.G. Proteins in action: The physics of structural fluctuations and conformational changes. Curr. Opin. Struct. Biol. 2003, 13, 552–557. [Google Scholar] [CrossRef] [PubMed]
- Gräter, F.; Grubmüller, H. Fluctuations of primary ubiquitin folding intermediates in a force clamp. J. Struct. Biol. 2007, 157, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Ptitsyn, O.B.; Creighton, T.E. Protein Folding; W.H. Freeman and Co.: New York, NY, USA, 1992; pp. 243–300. [Google Scholar]
- Samuel, D.; Kumar, T.; Srimathi, T.; Hsieh, H.c.; Yu, C. Identification and Characterization of an Equilibrium Intermediate in the Unfolding Pathway of an All β-Barrel Protein. J. Biol. Chem. 2000, 275, 34968–34975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobson, C.M.; Karplus, M. The fundamentals of protein folding: Bringing together theory and experiment. Curr. Opin. Struct. Biol. 1999, 9, 92–101. [Google Scholar] [CrossRef]
- Liu, Z.P.; Rizo, J.; Gierasch, L.M. Equilibrium folding studies of cellular retinoic acid binding protein, a predominantly beta-sheet protein. Biochemistry 1994, 33, 134–142. [Google Scholar] [CrossRef]
- Sivaraman, T.; Kumar, T.K.; Jayaraman, G.; Han, C.C.; Yu, C. Characterization of a partially structured state in an all-beta-sheet protein. Biochem. J. 1997, 321 Pt 2, 457–464. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, N.; Capaldi, A.P.; James, R.; Kleanthous, C.; Radford, S.E. Rapid folding with and without populated intermediates in the homologous four-helix proteins Im7 and Im9. J. Mol. Biol. 1999, 286, 1597–1608. [Google Scholar] [CrossRef]
- Bryngelson, J.D.; Onuchic, J.N.; Socci, N.D.; Wolynes, P.G. Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins 1995, 21, 167–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruebele, M. Downhill protein folding: Evolution meets physics. Comptes Rendus Biol. 2005, 328, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.M.; Sharpe, T.D. Protein Folding, Energy Landscapes and Downhill Protein Folding Scenarios. In Encyclopedia of Biophysics; Roberts, G., Watts, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 1–19. [Google Scholar]
- Judy, E.; Kishore, N. A look back at the molten globule state of proteins: Thermodynamic aspects. Biophys. Rev. 2019, 11, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Paci, E.; Smith, L.; Dobson, C.; Karplus, M. Exploration of partially unfolded states of human α-lactalbumin by molecular dynamics simulation. J. Mol. Biol. 2001, 306, 329–347. [Google Scholar] [CrossRef]
- Ikeguchi, M.; Kuwajima, K.; Mitani, M.; Sugai, S. Evidence for identity between the equilibrium unfolding intermediate and a transient folding intermediate: A comparative study of the folding reactions of alpha-lactalbumin and lysozyme. Biochemistry 1986, 25, 6965–6972. [Google Scholar] [CrossRef]
- Dasmeh, P.; Kepp, K.P. Unfolding simulations of holomyoglobin from four mammals: Identification of intermediates and beta-sheet formation from partially unfolded states. PLoS ONE 2013, 8, e80308. [Google Scholar] [CrossRef] [Green Version]
- Blum, O.; Haiek, A.; Cwikel, D.; Dori, Z.; Meade, T.J.; Gray, H.B. Isolation of a myoglobin molten globule by selective cobalt(III)-induced unfolding. Proc. Natl. Acad. Sci. USA 1998, 95, 6659–6662. [Google Scholar] [CrossRef] [Green Version]
- Cort, J.R.; Andersen, N.H. Formation of a Molten-Globule-like State of Myoglobin in Aqueous Hexafluoroisopropanol. Biochem. Biophys. Res. Commun. 1997, 233, 687–691. [Google Scholar] [CrossRef]
- Lin, L.; Pinker, R.J.; Forde, K.; Rose, G.D.; Kallenbach, N.R. Molten globular characteristics of the native state of apomyoglobin. Nat. Struct. Biol. 1994, 1, 447–452. [Google Scholar] [CrossRef]
- Barrick, D.; Baldwin, R.L. The molten globule intermediate of apomyoglobin and the process of protein folding. Protein Sci. 1993, 2, 869–876. [Google Scholar] [CrossRef]
- Fink, A.L.; Oberg, K.A.; Seshadri, S. Discrete intermediates versus molten globule models for protein folding: Characterization of partially folded intermediates of apomyoglobin. Fold. Des. 1998, 3, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Parray, Z.A.; Ahmad, F.; Alajmi, M.F.; Hussain, A.; Hassan, M.I.; Islam, A. Formation of molten globule state in horse heart cytochrome c under physiological conditions: Importance of soft interactions and spectroscopic approach in crowded milieu. Int. J. Biol. Macromol. 2020, 148, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.H.; Kumar, A.; Prakash, A.; Taneja, B.; Islam, A.; Hassan, M.I.; Ahmad, F. Structural and thermodynamic characterisation of L94F mutant of horse cytochrome c. Int. J. Biol. Macromol. 2016, 92, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Kamiyama, T.; Sadahide, Y.; Nogusa, Y.; Gekko, K. Polyol-induced molten globule of cytochrome c: An evidence for stabilization by hydrophobic interaction. Biochim. Biophys. Acta 1999, 1434, 44–57. [Google Scholar] [CrossRef]
- Hamada, D.; Kidokoro, S.; Fukada, H.; Takahashi, K.; Goto, Y. Salt-induced formation of the molten globule state of cytochrome c studied by isothermal titration calorimetry. Proc. Natl. Acad. Sci. USA 1994, 91, 10325–10329. [Google Scholar] [CrossRef] [Green Version]
- Dar, M.A.; Islam, A.; Hassan, M.I.; Ahmad, F. Effect of mammalian kidney osmolytes on the folding pathway of sheep serum albumin. Int. J. Biol. Macromol. 2017, 97, 625–634. [Google Scholar] [CrossRef]
- Samaddar, S.; Mandal, A.K.; Mondal, S.K.; Sahu, K.; Bhattacharyya, K.; Roy, S. Solvation Dynamics of a Protein in the Pre Molten Globule State. J. Phys. Chem. B 2006, 110, 21210–21215. [Google Scholar] [CrossRef]
- Zerovnik, E.; Jerala, R.; Kroon-Zitko, L.; Pain, R.H.; Turk, V. Intermediates in denaturation of a small globular protein, recombinant human stefin B. J. Biol. Chem. 1992, 267, 9041–9046. [Google Scholar] [CrossRef]
- Daggett, V.; Levitt, M. A model of the molten globule state from molecular dynamics simulations. Proc. Natl. Acad. Sci. USA 1992, 89, 5142–5146. [Google Scholar] [CrossRef] [Green Version]
- Farrell, H.M.; Qi, P.X.; Uversky, V.N. New Views of Protein Structure: Applications to the Caseins: Protein Structure and Functionality. In Advances in Biopolymers; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 2006; Volume 935, pp. 52–70. [Google Scholar]
- Morozova, L.A.; Haynie, D.T.; Arico-Muendel, C.; Dael, H.V.; Dobson, C.M. Structural basis of the stability of a lysozyme molten globule. Nat. Struct. Biol. 1995, 2, 871–875. [Google Scholar] [CrossRef]
- Robertson, A.D.; Baldwin, R.L. Hydrogen exchange in thermally denatured ribonuclease. Biochemistry 1991, 30, 9907–9914. [Google Scholar] [CrossRef] [PubMed]
- Harding, M.M.; Williams, D.H.; Woolfson, D.N. Characterization of a partially denatured state of a protein by two-dimensional NMR: Reduction of the hydrophobic interactions in ubiquitin. Biochemistry 1991, 30, 3120–3128. [Google Scholar] [CrossRef] [PubMed]
- García-Fandiño, R.; Bernadó, P.; Ayuso-Tejedor, S.; Sancho, J.; Orozco, M. Defining the Nature of Thermal Intermediate in 3 State Folding Proteins: Apoflavodoxin, a Study Case. PLoS Comput. Biol. 2012, 8, e1002647. [Google Scholar] [CrossRef] [PubMed]
- Hawe, A.; Sutter, M.; Jiskoot, W. Extrinsic fluorescent dyes as tools for protein characterization. Pharm. Res. 2008, 25, 1487–1499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uversky, V.N.; Karnoup, A.S.; Segel, D.J.; Seshadri, S.; Doniach, S.; Fink, A.L. Anion-induced folding of Staphylococcal nuclease: Characterization of multiple equilibrium partially folded intermediates. J. Mol. Biol. 1998, 278, 879–894. [Google Scholar] [CrossRef]
- Powers, M.V.; Workman, P. Inhibitors of the heat shock response: Biology and pharmacology. FEBS Lett. 2007, 581, 3758–3769. [Google Scholar] [CrossRef] [Green Version]
- Yu, A.; Fox, S.G.; Cavallini, A.; Kerridge, C.; O’Neill, M.J.; Wolak, J.; Bose, S.; Morimoto, R.I. Tau protein aggregates inhibit the protein-folding and vesicular trafficking arms of the cellular proteostasis network. J. Biol. Chem. 2019, 294, 7917–7930. [Google Scholar] [CrossRef]
- Saschenbrecker, S.; Bracher, A.; Rao, K.V.; Rao, B.V.; Hartl, F.U.; Hayer-Hartl, M. Structure and function of RbcX, an assembly chaperone for hexadecameric Rubisco. Cell 2007, 129, 1189–1200. [Google Scholar] [CrossRef] [Green Version]
- Shahnaj, S.; Potshangbam, A.M.; Chowhan, R.K.; Parray, Z.A.; Kakchingtabam, P.; Kumari, A.; Islam, A.; Khan, A.; Singh, L.R.; Rahaman, H. The anti-oxidant enzyme, Prdx6 might have cis-acting regulatory sequence(s). Int. J. Biol. Macromol. 2020, 149, 1139–1150. [Google Scholar] [CrossRef]
- Steffen, K.K.; Dillin, A. A Ribosomal Perspective on Proteostasis and Aging. Cell Metab. 2016, 23, 1004–1012. [Google Scholar] [CrossRef] [Green Version]
- Anisimova, A.S.; Alexandrov, A.I.; Makarova, N.E.; Gladyshev, V.N.; Dmitriev, S.E. Protein synthesis and quality control in aging. Aging 2018, 10, 4269–4288. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.H.; Shalini, S.; Filipovska, A.; Richman, T.R.; Davies, S.; Martin, S.D.; McGee, S.L.; Puccini, J.; Nikolic, A.; Dorstyn, L.; et al. Age-related proteostasis and metabolic alterations in Caspase-2-deficient mice. Cell Death Dis. 2015, 6, e1615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, T.; Eckhard, U.; Dufour, A.; Solis, N.; Overall, C.M. Proteolytic Cleavage-Mechanisms, Function, and ”Omic” Approaches for a Near-Ubiquitous Posttranslational Modification. Chem. Rev. 2018, 118, 1137–1168. [Google Scholar] [CrossRef] [PubMed]
- Bett, J.S. Proteostasis regulation by the ubiquitin system. Essays Biochem. 2016, 60, 143–151. [Google Scholar] [CrossRef]
- Wang, Y.; Lomakin, A.; Kanai, S.; Alex, R.; Benedek, G.B. Liquid–Liquid Phase Separation in Oligomeric Peptide Solutions. Langmuir 2017, 33, 7715–7721. [Google Scholar] [CrossRef]
- Remondelli, P.; Renna, M. The Endoplasmic Reticulum Unfolded Protein Response in Neurodegenerative Disorders and Its Potential Therapeutic Significance. Front. Mol. Neurosci. 2017, 10, 187. [Google Scholar] [CrossRef] [Green Version]
- Carra, S.; Brunsting, J.F.; Lambert, H.; Landry, J.; Kampinga, H.H. HspB8 participates in protein quality control by a non-chaperone-like mechanism that requires eIF2α phosphorylation. J. Biol. Chem. 2009, 284, 5523–5532. [Google Scholar] [CrossRef] [Green Version]
- Harada, R.; Inagaki, Y.; Shigeta, Y. Protein Folding and Evolution. In Reference Module in Materials Science and Materials Engineering; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Amara, J.F.; Cheng, S.H.; Smith, A.E. Intracellular protein trafficking defects in human disease. Trends Cell. Biol. 1992, 2, 145–149. [Google Scholar] [CrossRef]
- Neupert, W. Transport of proteins across mitochondrial membranes. Clin. Investig. 1994, 72, 251–261. [Google Scholar] [CrossRef] [Green Version]
- Marinko, J.T.; Huang, H.; Penn, W.D.; Capra, J.A.; Schlebach, J.P.; Sanders, C.R. Folding and Misfolding of Human Membrane Proteins in Health and Disease: From Single Molecules to Cellular Proteostasis. Chem. Rev. 2019, 119, 5537–5606. [Google Scholar] [CrossRef]
- Bhat, M.Y.; Dar, T.A.; Singh, L.R. Casein Proteins: Structural and Functional Aspects, Milk Proteins—From Structure to Biological Properties and Health Aspects; IntechOpen: London, UK, 2016. [Google Scholar] [CrossRef] [Green Version]
- Anfinsen, C.B. Principles that govern the folding of protein chains. Science 1973, 181, 223–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rose, G.D.; Fleming, P.J.; Banavar, J.R.; Maritan, A. A backbone-based theory of protein folding. Proc. Natl. Acad. Sci. USA 2006, 103, 16623–16633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grushevskyi, E.O.; Kukaj, T.; Schmauder, R.; Bock, A.; Zabel, U.; Schwabe, T.; Benndorf, K.; Lohse, M.J. Stepwise activation of a class C GPCR begins with millisecond dimer rearrangement. Proc. Natl. Acad. Sci. USA 2019, 116, 10150–10155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; He, X.; Yang, Z.; Chai, Z.; Zhou, S.; Wang, J.; Rehman, A.U.; Ni, D.; Pu, J.; Sun, J.; et al. Activation pathway of a G protein-coupled receptor uncovers conformational intermediates as targets for allosteric drug design. Nat. Commun. 2021, 12, 4721. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, Y.; Ni, D.; Huang, Z.; Wei, J.; Feng, L.; Su, J.C.; Wei, Y.; Ning, S.; Yang, X.; et al. Targeting a cryptic allosteric site of SIRT6 with small-molecule inhibitors that inhibit the migration of pancreatic cancer cells. Acta Pharm. Sin. B 2022, 12, 876–889. [Google Scholar] [CrossRef]
- Lu, S.; Ni, D.; Wang, C.; He, X.; Lin, H.; Wang, Z.; Zhang, J. Deactivation Pathway of Ras GTPase Underlies Conformational Substates as Targets for Drug Design. ACS Catal. 2019, 9, 7188–7196. [Google Scholar] [CrossRef]
- Ni, D.; Wei, J.; He, X.; Rehman, A.U.; Li, X.; Qiu, Y.; Pu, J.; Lu, S.; Zhang, J. Discovery of cryptic allosteric sites using reversed allosteric communication by a combined computational and experimental strategy. Chem. Sci. 2021, 12, 464–476. [Google Scholar] [CrossRef]
- Shukla, D.; Meng, Y.; Roux, B.; Pande, V. Activation pathway of Src kinase reveals intermediate states as targets for drug design. Nat. Commun. 2014, 5, 3397. [Google Scholar] [CrossRef]
- Oruganty, K.; Talathi, N.S.; Wood, Z.A.; Kannan, N. Identification of a hidden strain switch provides clues to an ancient structural mechanism in protein kinases. Proc. Natl. Acad. Sci. USA 2013, 110, 924–929. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.-I.; Voegtli, W.C.; Sturgis, H.L.; Dizon, F.P.; Vigers, G.P.A.; Brandhuber, B.J. Crystal Structure of Human AKT1 with an Allosteric Inhibitor Reveals a New Mode of Kinase Inhibition. PLoS ONE 2010, 5, e12913. [Google Scholar] [CrossRef]
- Lundvig, D.; Lindersson, E.; Jensen, P.H. Pathogenic effects of alpha-synuclein aggregation. Mol. Brain Res. 2005, 134, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. A protein-chameleon: Conformational plasticity of alpha-synuclein, a disordered protein involved in neurodegenerative disorders. J. Biomol. Struct. Dyn. 2003, 21, 211–234. [Google Scholar] [CrossRef] [PubMed]
- Neudecker, P.; Robustelli, P.; Cavalli, A.; Walsh, P.; Lundström, P.; Zarrine-Afsar, A.; Sharpe, S.; Vendruscolo, M.; Kay, L.E. Structure of an intermediate state in protein folding and aggregation. Science 2012, 336, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Webster, J.M.; Darling, A.L.; Uversky, V.N.; Blair, L.J. Small Heat Shock Proteins, Big Impact on Protein Aggregation in Neurodegenerative Disease. Front. Pharmacol. 2019, 10, 1047. [Google Scholar] [CrossRef]
- Feige, M.J.; Groscurth, S.; Marcinowski, M.; Yew, Z.T.; Truffault, V.; Paci, E.; Kessler, H.; Buchner, J. The structure of a folding intermediate provides insight into differences in immunoglobulin amyloidogenicity. Proc. Natl. Acad. Sci. USA 2008, 105, 13373–13378. [Google Scholar] [CrossRef] [Green Version]
- Reynaud, E. Protein Misfolding and Degenerative Diseases. Nat. Educ. 2010, 3, 28. [Google Scholar]
- Luthra, P.M. Paradigm of protein folding in neurodegenerative diseases. Adv. Proteom. Res. 2012, 2, 223–252. [Google Scholar]
- Thorn, D.C.; Ecroyd, H.; Carver, J.A. Chapter 30—Polymorphism in Casein Protein Aggregation and Amyloid Fibril Formation. In Bio-Nanoimaging; Uversky, V.N., Lyubchenko, Y.L., Eds.; Academic Press: Boston, MA, USA, 2014; pp. 323–331. [Google Scholar]
- Głąb, T.K.; Boratyński, J. Potential of Casein as a Carrier for Biologically Active Agents. Top. Curr. Chem. 2017, 375, 71. [Google Scholar] [CrossRef] [Green Version]
- Thorn, D.C.; Ecroyd, H.; Carver, J.A.; Holt, C. Casein structures in the context of unfolded proteins. Int. Dairy J. 2015, 46, 2–11. [Google Scholar] [CrossRef] [Green Version]
- Bychkova, V.; Ptitsyn, O. Folding intermediates are involved in genetic diseases? FEBS Lett. 1995, 359, 6–8. [Google Scholar] [CrossRef] [Green Version]
- Clancy, S. Genetic mutation. Nat. Educ. 2008, 1, 187. [Google Scholar]
- Bychkova, V.E.; Ptitsyn, O.B. The state of unfolded globules of protein molecules is more quickly becoming a rule, rather than an exception. Biofizika 1993, 38, 58–66. [Google Scholar] [PubMed]
- Buchanan, S.K. β-barrel proteins from bacterial outer membranes: Structure, function and refolding. Curr. Opin. Struct. Biol. 1999, 9, 455–461. [Google Scholar] [CrossRef]
- Bychkova, V.E.; Pain, R.H.; Ptitsyn, O.B. The ‘molten globule’ state is involved in the translocation of proteins across membranes? FEBS Lett. 1988, 238, 231–234. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.; Rye, H.S. GroEL-mediated protein folding: Making the impossible, possible. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 211–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinker, A.; Pfeifer, G.; Kerner, M.J.; Naylor, D.J.; Hartl, F.U.; Hayer-Hartl, M. Dual Function of Protein Confinement in Chaperonin-Assisted Protein Folding. Cell 2001, 107, 223–233. [Google Scholar] [CrossRef] [Green Version]
- Hayer-Hartl, M. From chaperonins to Rubisco assembly and metabolic repair. Protein Sci. 2017, 26, 2324–2333. [Google Scholar] [CrossRef]
- Fonin, A.V.; Darling, A.L.; Kuznetsova, I.M.; Turoverov, K.K.; Uversky, V.N. Intrinsically disordered proteins in crowded milieu: When chaos prevails within the cellular gumbo. Cell. Mol. Life Sci. CMLS 2018, 75, 3907–3929. [Google Scholar] [CrossRef]
- Kachel, N.; Kremer, W.; Zahn, R.; Kalbitzer, H.R. Observation of intermediate states of the human prion protein by high pressure NMR spectroscopy. BMC Struct. Biol. 2006, 6, 16. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parray, Z.A.; Shahid, M.; Islam, A. Insights into Fluctuations of Structure of Proteins: Significance of Intermediary States in Regulating Biological Functions. Polymers 2022, 14, 1539. https://doi.org/10.3390/polym14081539
Parray ZA, Shahid M, Islam A. Insights into Fluctuations of Structure of Proteins: Significance of Intermediary States in Regulating Biological Functions. Polymers. 2022; 14(8):1539. https://doi.org/10.3390/polym14081539
Chicago/Turabian StyleParray, Zahoor Ahmad, Mohammad Shahid, and Asimul Islam. 2022. "Insights into Fluctuations of Structure of Proteins: Significance of Intermediary States in Regulating Biological Functions" Polymers 14, no. 8: 1539. https://doi.org/10.3390/polym14081539