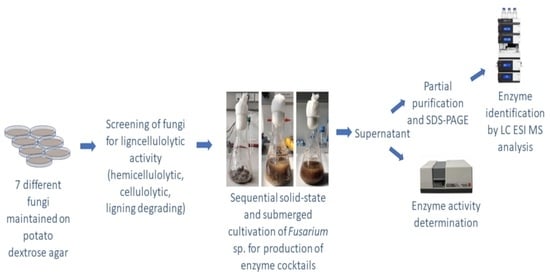
Screening of Lignocellulolytic Enzyme Activities in Fungal Species and Sequential Solid-State and Submerged Cultivation for the Production of Enzyme Cocktails
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Setup
2.2. Feedstock
2.3. Working Microorganisms and Media
2.4. Cellulolytic Enzyme Activity Plate Test
2.5. Preculture Cultivation for the Induction of Lignocellulolitic Enzyme Production
2.6. Lignocellulolytic Enzyme Production
2.7. Protein Purification
2.8. Analytical Methods
2.8.1. Cellulolytic and Hemicellulolytic Activity Assay
2.8.2. Laccase Activity Assay
2.8.3. Lignin Peroxidase (LiP) Assay
2.8.4. Cellobiose Dehydrogenase Assay
2.8.5. Bradford Protein Assay
2.8.6. Sodium Dodecyl Sulphate Polyacrylamide Gel Electrophoresis (SDS-PAGE)
2.8.7. Peptide Profiling by LC-ESI-MS
2.8.8. Statistical Analysis
3. Results and Discussion
3.1. Screening of Fungi for Lignocellulolytic Enzyme Activity
3.1.1. Hemicellulolytic Activity
3.1.2. Cellulolytic Activity
3.1.3. Lignin Degrading Activity
3.2. Sequential Solid State and Submerged Cultivation of Selected Fungi Strains
Partial Purification and LC-ESI-MS Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bušić, A.; Marđetko, N.; Kundas, S.; Morzak, G.; Belskaya, H.; Ivančić Šantek, M.; Komes, D.; Novak, S.; Šantek, B. Bioethanol Production from Renewable Raw Materials and Its Separation and Purification: A Review. Food Technol. Biotech. 2018, 56, 289–311. [Google Scholar] [CrossRef] [PubMed]
- Kostylev, M.; Wilson, D. Synergistic interactions in cellulose hydrolysis. Biofuels 2012, 3, 61–70. [Google Scholar] [CrossRef]
- Andlar, M.; Rezić, T.; Marđetko, N.; Kracher, D.; Ludwig, R.; Šantek, B. Lignocellulose degradation: An overview of fungi and fungal enzymes involved in lignocellulose degradation. Eng. Life Sci. 2018, 18, 768–778. [Google Scholar] [CrossRef]
- Van Dyk, J.S.; Pletschke, B.I. A review of lignocellulose bioconversion using enzymatic hydrolysis and synergistic cooperation between enzymes-Factors affecting enzymes, conversion and synergy. Biotech. Adv. 2012, 30, 1458–1480. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, N.; Rawat, R.; Oberoi, H.S.; Ramteke, P.W. A review on fuel ethanol production from lignocellulosic biomass. Int. J. Green Energy 2015, 12, 949–960. [Google Scholar] [CrossRef]
- Haq, F.; Ali, H.; Shuaib, M.; Badshah, M.; Hassan, S.W.; Munis, M.F.H.; Chaudhary, H.J. Recent progress in bioethanol production from lignocellulosic materials: A review. Int. J. Green. Energy 2016, 13, 1413–1441. [Google Scholar] [CrossRef]
- Motta, F.A.C.; Santana, M. A review of xylanase production by the fermentation of xylan: Classification, characterization and applications. In Sustainable Degradation of Lignocellulosic Biomass—Techniques, Applications and Commercialization; Chandel, A.K., da Silva, S.S., Eds.; InTech: Rijeka, Croatia, 2013. [Google Scholar]
- Dashtban, M.; Schraft, H.; Syed, T.A.; Qin, W. Fungal biodegradation and enzymatic modification of lignin. Int. J. Biochem. Mol. Biol. 2010, 1, 36–50. [Google Scholar]
- Farias, N.; Almeida, I.; Meneses, C. New Bacterial Phytase through Metagenomic Prospection. Molecules 2018, 23, 448. [Google Scholar] [CrossRef] [Green Version]
- Acharya, D.K.; Chabhadiya, S.B.; Shah, A.J.; Shilpkar, P.; Acharya, P.B.; Modi, H.A. Enzyme profiling of lignocellulolytic fungi. Int. J. Biol. Chem. Sci. 2010, 4, 443–449. [Google Scholar] [CrossRef]
- Irbe, I.; Elisashvili, V.; Asatiani, M.; Janberga, A.; Andersone, I.; Andersons, B.; Biziks, V.; Grinins, J. Lignocellulolytic activity of Coniophora puteana and Trametes versicolor in fermentation of wheat bran and decay of hydrothermally modified hardwoods. Int. Biodeter. Biodegr. 2014, 86, 71–78. [Google Scholar] [CrossRef]
- Sajith, S.; Priji, P.; Sreedevi, S.; Benjamin, S. An overwiev on fungal cellulases with an industrial perspective. J. Nutr. Food. Sci. 2016, 6, 461. [Google Scholar] [CrossRef] [Green Version]
- Papagianni, M. Fungal morphology and metabolite production in submerged mycelial processes. Biotechnol. Adv. 2004, 22, 189–259. [Google Scholar] [CrossRef] [PubMed]
- Domingues, F.C.; Queiroz, J.A.; Cabral, J.M.; Fonseca, L.P. The influence of culture conditions on mycelial structure and cellulase production by Trichoderma reesei Rut C-30. Enzyme Microb. Technol. 2000, 26, 394–401. [Google Scholar] [CrossRef]
- An, Q.; Wu, X.J.; Han, M.L.; Cui, B.K.; He, S.H.; Dai, Y.C.; Si, J. Sequential solid-state and submerged cultivation of the white rot fungus Pleurotus ostreatus on biomass and the activity of lignocellulolytic enzymes. BioResources 2016, 11, 8791–8805. [Google Scholar] [CrossRef] [Green Version]
- Sohail, M.; Siddiqi, R.; Ahmad, A.; Khan, S.A. Cellulase production from Aspergillus niger MS82: Effect of temperature and pH. New Biotechnol. 2009, 25, 437–441. [Google Scholar] [CrossRef]
- Ahamed, A.; Vermette, P. Culture-based strategies to enhance cellulase enzyme production from Trichoderma reesei RUT-C30 in bioreactor culture conditions. Biochem. Eng. J. 2008, 40, 399–407. [Google Scholar] [CrossRef]
- Castro, A.M.; Albuquerque De Carvalho, M.L.; Leite, S.G.F.; Pereira, N., Jr. Cellulases from Penicillium funiculosum: Production, properties and application to cellulose hydrolysis. J. Ind. Microbiol. Biotechnol. 2010, 37, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Maeda, R.N.; Serpa, V.I.; Rocha, V.A.L.; Mesquita, R.A.A.; De Santa Anna, L.M.M.; Castro, A.M.; Driemeier, C.E.; Pereira, N., Jr.; Polikarpov, I. Enzymatic hydrolysis of pretreated sugar cane bagasse using Penicillium funiculosum and Trichoderma harzianum cellulases. Process Biochem. 2011, 46, 1196–1201. [Google Scholar] [CrossRef]
- Singhania, R.R.; Sukumaran, R.K.; Patel, A.K. Advancement and comparative profiles in the production technologies using solid-state and submerged fermentation for microbial cellulases. Enzyme Microb. Technol. 2010, 46, 541–549. [Google Scholar] [CrossRef]
- Farinas, C.S.; Vitcosque, G.L.; Fonseca, R.F.; Neto, V.B.; Couri, S. Modeling the effects of solid state fermentation operating conditions on endoglucanase production using an instrumented bioreactor. Ind. Crop. Prod. 2011, 34, 1186–1192. [Google Scholar] [CrossRef]
- Cunha, F.M.; Esperança, M.N.; Zangirolami, T.C.; Badino, A.C.; Farinas, C.S. Sequential solid-state and submerged cultivation of Aspergillus niger on sugarcane bagasse for the production of cellulase. Bioresour. Technol. 2012, 112, 270–274. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Zhou, H.; Li, D.; Zhang, H.; Wang, H.; Lu, F. Optimized expression and enhanced production of alkaline protease by genetically modified Bacillus licheniformis 2709. Microb. Cell Factories 2020, 19, 45. [Google Scholar] [CrossRef] [PubMed]
- Polizeli, M.; Rizzatti, A.C.S.; Monti, R. Xylanases from fungi: Properties and industrial applications. Appl. Microbiol. Biotechnol. 2005, 67, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Marđetko, N.; Novak, M.; Trontel, A.; Grubišić, M.; Galić, M.; Šantek, B. Bioethanol production from dilute-acid pre-treated wheat straw liquor hydrolysate by genetically engineered Saccharomy-ces cerevisiae. Chem. Biochem. Eng. Q. 2018, 32, 483–499. [Google Scholar] [CrossRef]
- Sluiter, A.; Hames, B.; Ruiz, R.; Scarlata, C.; Sluiter, J.; Templeton, D.; Crocker, D. Determination of structural carbohydrates and lignin in biomass. NREL Lab. Anal. Proced. 2012, 1617, 1–6. [Google Scholar]
- Pečiulytė, D. Isolation of cellulolytic fungi from waste paper gradual recycling materials. Ekologija 2007, 53, 11–18. [Google Scholar]
- Florencio, C.; Couri, S.; Sanchez Farinas, C. Correlation between agar plate screening and solid-state fermentation for the prediction of cellulase production by Trichoderma strains. Enzyme Res. 2012, 2012, 793708. [Google Scholar] [CrossRef] [Green Version]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Ghose, T.K. Measurement of cellulase activities. Pure Appl. Chem. 1987, 59, 257–268. [Google Scholar] [CrossRef]
- More, S.S.; Renuka, P.S.; Pruthvi, K.; Swetha, M.; Malini, S.; Veena, S.M. Isolation, Purification, and Characterization of Fungal Laccase from Pleurotus sp. Enzyme Res. 2011, 2011, 248735. [Google Scholar] [CrossRef] [Green Version]
- Daljit, S.A.; Paramjit, K.G. Comparison of two assay procedures for lignin peroxidase. Enzyme Microb. Technol. 2001, 28, 602–605. [Google Scholar] [CrossRef]
- Ma, S.; Preims, M.; Piumi, F.; Kappel, L.; Seiboth, B.; Record, E.; Kracher, D.; Ludwig, R. Molecular and catalytic properties of fungal extracellular cellobiose dehydrogenase produced in prokaryotic and eukaryotic expression systems. Microb. Cell. Fact. 2017, 16, 37. [Google Scholar] [CrossRef] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Halada, P.; Leitner, C.; Sedmera, P.; Haltrich, D.; Volc, J. Identification of the covalent flavin adenine dinucleotide-binding region in pyranose 2-oxidase from Trametes multicolor. Anal. Biochem. 2003, 314, 235–242. [Google Scholar] [CrossRef]
- M’Barek, H.N.; Taidi, B.; Smaoui, T.; Ben Aziz, M.; Mansouri, A. Isolation, screening and identification of ligno-cellulolytic fungi from northern central Morocco. Biotechnol. Agron. Soc. Environ. 2019, 23, 207–217. [Google Scholar] [CrossRef]
- Saroj, P.P.M.; Narasimhulu, K. Characterization of thermophilic fungi producing extracellular lignocellulolytic enzymes for lignocellulosic hydrolysis under solid-state fermentation. Bioresour. Bioprocess. 2018, 5, 31. [Google Scholar] [CrossRef] [Green Version]
- Elisashvili, V.; Kachlishvili, E.; Penninckx, M.J.; Tsiklauri, N.; Metreveli, E.; Kharziani, T.; Giorgi Kvesitadze, G. Lentinus edodes and Pleurotus species lignocellulolytic enzymes activity in submerged and solid-state fermentation of lignocellulosic wastes of different composition. Biores. Technol. 2008, 99, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Isikhuemhen, O.S.; Mikiashvilli, N.A. Lignocellulolytic enzyme activity, substrate utilization, and mushroom yield by Pleurotus ostreatus cultivated on substrate containing anaerobic digester solids. J. Ind. Microbiol. Biotechnol. 2009, 36, 1353–1362. [Google Scholar] [CrossRef] [PubMed]
- Lemos, J.L.S.; de Fontes, M.C.A.; Pereira, N. Xylanase Production by Aspergillus awamori in Solid-State Fermentation and Influence of Different Nitrogen Sources. In Twenty-Second Symposium on Biotechnology for Fuels and Chemicals; Davison, B.H., McMillan, J., Finkelstein, M., Eds.; ABAB Symposium; Humana Press: Totowa, NJ, USA, 2001. [Google Scholar] [CrossRef]
- Yang, S.Q.; Yan, Q.J.; Jiang, Z.Q.; Li, L.T.; Tian, H.M.; Wang, Y.Z. High-level of xylanase production by the thermophilic Paecilomyces themophila J18 on wheat straw in solid-state fermentation. Biores. Technol. 2006, 97, 1794–1800. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.G.; Sindhu, R.; Shashidhar, S. Fungal xylanase production under solid state and submerged fermentation conditions. Afr. J. Microbiol. Res. 2008, 2, 82–86. [Google Scholar] [CrossRef]
- Simones, M.L.G.; Tauk-Tornisielo, S.M.; Tapia, D.M. Screening of culture conditions for xylanase production by filamentous fungi. Afr. J. Biotechnol. 2009, 8, 6317–6326. [Google Scholar] [CrossRef]
- Bakri, Y.; Masson, M.; Thonart, P. Isolation and identification of two new fungal strains for xylanase production. Appl. Biochem. Biotechnol. 2010, 162, 1626–1634. [Google Scholar] [CrossRef]
- Knob, A.; Fortkamp, D.; Prolo, T.; Izidoro, S.C.; Almeida, J.M. Agro-residues as alternative for xylanase production by filamentous fungi. BioResources 2014, 9, 5738–5773. [Google Scholar] [CrossRef]
- Saha, B. Xylanase from a newly isolated Fusarium verticillioides capable of utilizing corn fiber xylan. Appl. Microbiol. Biotechnol. 2001, 56, 762–766. [Google Scholar] [CrossRef]
- Arabi, M.I.; Bakri, Y.; Jawhar, M. Extracellular xylanase production by Fusarium species in solid state fermentation. Pol. J. Microbiol. 2011, 60, 209–212. [Google Scholar] [CrossRef]
- Ögel, Z.B.; Yarangümeli, K.; Dündar, H.; Ifrij, İ. Submerged cultivation of scytalidium thermophilum on complex lignocellulosic biomass for endoglucanase production. Enzyme Microb. Technol. 2001, 28, 689–695. [Google Scholar] [CrossRef]
- Shu, C.H.; Xu, C.J.; Lin, E.S. Production, purification and partial characterization of a novel endo-β-1,3-glucanase from Agaricus brasiliensis. Process Biochem. 2006, 41, 1229–1233. [Google Scholar] [CrossRef]
- Martin, K.; McDougall, B.M.; McIlroy, S.; Chen, J.J.; Seviour, R.J. Biochemistry and molecular biology of exocellular fungal β-(1,3)- and β-(1,6)-glucanases. FEMS Microbiol. Rev. 2007, 31, 168–192. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Qiaojuan, Y.; Jiang, Z.; Fan, G.; Wang, L. Biochemical characterization of a novel thermostable β-1,3-1,4-Glucanase (Lichenase) from Paecilomyces thermophila. J. Agri. Food Chem. 2008, 56, 5345–5351. [Google Scholar] [CrossRef]
- Vinche, M.H.; Khanahmadi, M.; Ataei, S.A.; Danafar, F. Optimization of Process Variables for Production of Beta-Glucanase by Aspergillus niger CCUG33991 in Solid-State Fermentation Using Wheat Bran. Waste Biomass Valor. 2021, 12, 3233–3243. [Google Scholar] [CrossRef]
- Ravalason, H.; Grisel, S.; Chevret, D.; Favel, A.; Berrin, J.G. Fusarium verticillioides secretome as a source of auxiliary enzymes to enhance saccharifcation of wheat straw. Bioresour. Technol. 2012, 114, 589–596. [Google Scholar] [CrossRef]
- deb Dutta, S.; Tarafder, M.; Islam, R.; Datta, B. Characterization of cellulolytic enzymes of Fusarium soil isolates. Biocatal. Agric. Biotechnol. 2018, 14, 279–285. [Google Scholar] [CrossRef]
- Berka, R.M.; Grigoriev, I.V.; Otillar, R.; Salamov, A.; Grimwood, J.; Reid, I.; Ishmael, N.; John, T.; Darmond, C.; Moisan, M.C.; et al. Comperative genomic analysis of the thermophilic biomass degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat. Biotechnol. 2011, 29, 922–927. [Google Scholar] [CrossRef]
- Margaritis, A.; Merchant, R.F. Optimization of fermentation conditions for thermostable cellulase production by Thielavia terrestris. J. Ind. Microbiol. 1986, 1, 149–156. [Google Scholar] [CrossRef]
- Herrera Bravo de Laguna, I.; Toledo Marante, F.J.; Mioso, R. Enzymes and bioproducts produced by the ascomycete fungus Paecilomyces variotii. J. Appl. Microbiol. 2015, 119, 1455–1466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janusz, G.; Pawlik, A.; Sulej, J.; Świderska-Burek, U.; Jarosz-Wilkołazka, A.; Paszczyński, A. Lignin degradation: Microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 2017, 41, 941–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bugg, T.D.H.; Ahmad, M.; Hardiman, E.M.; Rahmanpour, R. Pathways for degredation of lignin in bacteria and fungi. Nat. Prod. Rep. 2011, 12, 1883–1896. [Google Scholar] [CrossRef]
- Bentil, J.A.; Anders Thygesen, A.; Moses Mensah, M.; Lange, L.; Meyer, A.S. Cellulase production by white-rot basidiomycetous fungi: Solid-state versus submerged cultivation. Appl. Microbiol. Biotechnol. 2018, 102, 5827–5839. [Google Scholar] [CrossRef]
- Pleissner, D.; Him Kwan, T.; Sze Ki Lin, C. Fungal hydrolysis in submerged fermentation for food waste treatment and fermentation feedstock preparation. Bioresour. Technol. 2014, 158, 48–54. [Google Scholar] [CrossRef]
- Martău, G.-A.; Unger, P.; Schneider, R.; Venus, J.; Vodnar, D.C.; López-Gómez, J.P. Integration of Solid State and Submerged Fermentations for the Valorization of Organic Municipal Solid Waste. J. Fungi 2021, 7, 766. [Google Scholar] [CrossRef] [PubMed]
- López-Gómez, J.P.; Venus, J. Potential Role of Sequential Solid-State and Submerged-Liquid Fermentations in a Circular Bioeconomy. Fermentation 2021, 7, 76. [Google Scholar] [CrossRef]
- Brito Codato, C.; Gaspar Bastos, R.; Ceccato-Antonini, S.R. Sequential process of solid-state cultivation with fungal consortium and ethanol fermentation by Saccharomyces cerevisiae from sugarcane bagasse. Bioprocess Biosyst. Eng. 2021, 44, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Elegbede, J.A.; Lateef, A. Valorization of Corn-Cob by Fungal Isolates for Production of Xylanase in Submerged and Solid State Fermentation Media and Potential Biotechnological Applications. Waste Biomass Valor. 2018, 9, 1273–1287. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L.; Chen, H. Correlations of medium physical properties and process performance in solid-state fermentation. Chem. Eng. Sci. 2017, 165, 65–73. [Google Scholar] [CrossRef]
- da Silva Menezes, B.; Misturini Rossi, D.; Záchia Ayub, M.A. Screening of filamentous fungi to produce xylanase and xylooligosaccharides in submerged and solid-state cultivations on rice husk, soybean hull, and spent malt as substrates. World J. Microbiol. Biotechnol. 2017, 33, 58. [Google Scholar] [CrossRef]
- Matrawy, A.A.; Khalil, A.I.; Marey, H.S.; Embaby, A.M. Use of Wheat Straw for Value-Added Product Xylanase by Penicillium chrysogenum Strain A3 DSM105774. J. Fungi 2021, 7, 696. [Google Scholar] [CrossRef] [PubMed]
- Köhl, J.; Lombaers, C.; Moretti, A.; Bandyopadhyay, R.; Somma, S.; Kastelein, P. Analysis of microbial taxonomical groups present in maize stalks suppressive to colonization by toxigenic Fusarium spp.: A strategy for the identification of potential antagonists. Biol. Control 2015, 83, 20–28. [Google Scholar] [CrossRef]
- Palumbo, J.D.; O’Keeffe, T.L.; Abbas, H.K. Microbial interactions with mycotoxigenic fungi and mycotoxins. Toxin Rev. 2008, 27, 261–285. [Google Scholar] [CrossRef]
- Meena, M.; Swapnil, P.; Zehra, A.; Kumar Dubey, M.; Upadhyay, R.S. Antagonistic assessment of Trichoderma spp. by producing volatile and non-volatile compounds against different fungal pathogens. Arch. Phytopathol. Pflanzenschutz 2017, 50, 629–648. [Google Scholar] [CrossRef]



| Component (g L−1) | Nutrient Medium | |||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| Yeast extract | 2 | - | 2 | - |
| Peptone | 5 | 2 | 5 | - |
| Corn steep liquor | - | 5 | - | 5 |
| Diammonium phosphate | 1.4 | 1.4 | 1.4 | 1.4 |
| KH2PO4 | 2 | 2 | 2 | 2 |
| MgSO4 | 0.2 | 0.2 | 0.2 | 0.2 |
| Urea | - | - | 0.3 | - |
| CaCl2 | - | - | 0.2 | - |
| Tween 80 | - | - | 1 | - |
| MnSO4 | - | - | - | 0.01 |
| CuSO4 | - | - | - | 0.01 |
| NaCl | - | - | - | 0.1 |
| Salt solution * (%) | 0.1 | - | 0.1 | - |
| Common Name | Substrate | Released Sugar |
|---|---|---|
| Starch degrading | Starch | D-glucose |
| Xylanase | Xylan | D-xylose |
| Mannanase | Mannan | D-mannose |
| Arabinase | Arabinan | L-arabinose |
| Endoglucanase | Cellulose | D-glucose |
| Exoglucanase | Carboxymethyl cellulose | D-glucose |
| Pectinase | Pectin | Galacturonic acid |
| Protein Concentration [mg mL−1] | Total Volume [mL] | Enzyme Activity [U mg−1total protein] | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Starch Degrading | Endo- and Exo-arabinase | Mannanase | Xylanase | Endoglucanase | Exoglucanase | Pectinase | |||
| FV crude extract | 0.41 ± 0.03 | 980 ± 23 | 0.42 ± 0.08 | 0.58 ± 0.07 | 0.53 ± 0.06 | 14.48 ± 0.12 | 0.21 ± 0.01 | 0.57 ± 0.01 | 1.022 ± 0.08 |
| FV partially purified extract | 0.89 ± 0.09 | 132 ± 8.4 | 0.91 ± 0.09 | 1.11 ± 0.10 | 1.75 ± 0.10 | 25.12 ± 0.25 | 0.55 ± 0.09 | 0.91 ± 0.07 | 4.45 ± 0.11 |
| FO crude extract | 0.33 ± 0.02 | 965 ± 37 | 0.76 ± 0.07 | 0.36 ± 0.04 | 0.62 ± 0.05 | 6.44 ± 0.18 | 0.34 ± 0.02 | 4.09 ± 0.01 | 0.79 ± 0.09 |
| FO partially purified extract | 0.61 ± 0.05 | 120 ± 5.6 | 1.09 ± 0.09 | 0.94 ± 0.07 | 1.47 ± 0.10 | 11.05 ± 0.14 | 1.09 ± 0.05 | 7.85 ± 0.18 | 2.93 ± 0.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marđetko, N.; Trontel, A.; Novak, M.; Pavlečić, M.; Ljubas, B.D.; Grubišić, M.; Tominac, V.P.; Ludwig, R.; Šantek, B. Screening of Lignocellulolytic Enzyme Activities in Fungal Species and Sequential Solid-State and Submerged Cultivation for the Production of Enzyme Cocktails. Polymers 2021, 13, 3736. https://doi.org/10.3390/polym13213736
Marđetko N, Trontel A, Novak M, Pavlečić M, Ljubas BD, Grubišić M, Tominac VP, Ludwig R, Šantek B. Screening of Lignocellulolytic Enzyme Activities in Fungal Species and Sequential Solid-State and Submerged Cultivation for the Production of Enzyme Cocktails. Polymers. 2021; 13(21):3736. https://doi.org/10.3390/polym13213736
Chicago/Turabian StyleMarđetko, Nenad, Antonija Trontel, Mario Novak, Mladen Pavlečić, Blanka Didak Ljubas, Marina Grubišić, Vlatka Petravić Tominac, Roland Ludwig, and Božidar Šantek. 2021. "Screening of Lignocellulolytic Enzyme Activities in Fungal Species and Sequential Solid-State and Submerged Cultivation for the Production of Enzyme Cocktails" Polymers 13, no. 21: 3736. https://doi.org/10.3390/polym13213736