Polycaprolactone/Polyethylene Glycol Blended with Dipsacus asper Wall Extract Nanofibers Promote Osteogenic Differentiation of Periodontal Ligament Stem Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of PDLSCs
2.2. Preparation the Extracts of DA
2.3. Fabrication of PCL/PEO Nanofibers Containing DAE
2.4. Characterization of Nanofiber
2.5. Cellular Viability vs. DAE Treatment
2.6. Calcium Quantification
2.7. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
2.8. Western Blotting
2.9. Statistical Analysis
3. Results
3.1. Characterization of PDLSCs
3.2. Optimization of Microwave-Assisted DAE Extraction
3.3. DAE Induces Osteogenic Differentiation
3.4. Osteoblastic Gene Expression
3.5. Calcium Quantification by OCN Staining
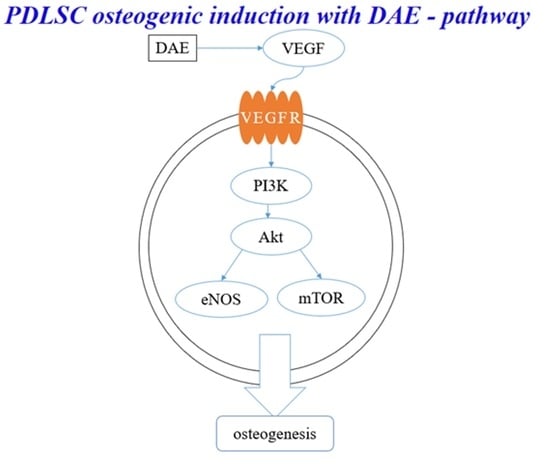
3.6. PI3K Pathway Assay
3.7. Optimization of Electrospinning
3.8. Characterization of Nanofibers
3.9. Nanofibers Containing DAE Induce Osteogenic Differentiation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tian, X.Y.; Wang, Y.H.; Liu, H.Y.; Yu, S.S.; Fang, W.S. On the Chemical Constituents of Dipsacus asper. Chem. Pharm. Bull. 2007, 55, 1677–1681. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Wei, B.; Peng, Z.; Fu, Q.; Wang, C.; Zhen, J.; Sun, J. Protective effects of Dipsacus asper polysaccharide on osteoporosis in vivo by regulating RANKL/RANK/OPG/VEGF and PI3K/Akt/eNOS pathway. Int. J. Biol. Macromol. 2019, 129, 579–587. [Google Scholar] [CrossRef]
- Chen, J.; Yao, D.; Yuan, H.; Zhang, S.; Tian, J.; Guo, W.; Liang, W.; Li, H.; Zhang, Y. Dipsacus asperoides polysaccharide induces apoptosis in osteosarcoma cells by modulating the PI3K/Akt pathway. Carbohydr. Polym. 2013, 95, 780–784. [Google Scholar] [CrossRef]
- Niu, Y.; Li, Y.; Huang, H.; Kong, X.; Zhang, R.; Liu, L.; Sun, Y.; Wang, T.; Mei, Q. Asperosaponin VI, A Saponin Component from Dipsacus asper Wall, induces Osteoblast Differentiation through Bone Morphogenetic Protein-2/p38 and Extracellular Signal-regulated Kinase 1/2 Pathway. Phytother. Res. 2011, 25, 1700–1706. [Google Scholar] [CrossRef]
- Niu, Y.B.; Kong, X.H.; Li, Y.H.; Fan, L.; Pan, Y.L.; Li, C.R.; Wu, X.L.; Lu, T.L.; Mei, Q.B. Radix Dipsaci total saponins stimulate MC3T3-E1 cell differentiation via the bone morphogenetic protein-2/MAPK/Smad-dependent Runx2 pathway. Mol. Med. Rep. 2015, 11, 4468–4472. [Google Scholar] [CrossRef] [PubMed]
- Remya, K.R.; Chandran, S.; Mani, S.; John, A.; Ramesh, P. Hybrid polycaprolactone/polyethylene oxide scaffolds with tunable fiber surface morphology, improved hydrophilicity and biodegradability for bone tissue engineering applications. J. Biomater. Sci. Polym. Ed. 2018, 29, 1444–1462. [Google Scholar] [CrossRef] [PubMed]
- Ates, B.; Koytepe, S.; Ulu, A.; Gurses, C.; Thakur, V.K. Chemistry, Structures, and Advanced Applications of Nanocomposites from Biorenewable Resources. Chem. Rev. 2020, 120, 9304. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.Y.; Su, W.T.; Chen, P.H. Comparing the effects of chitosan scaffolds containing various divalent metal phosphates on osteogenic differentiation of stem cells from human exfoliated deciduous teeth. Biol. Trace Elem. Res. 2018, 185, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Su, W.T.; Pan, Y.J. Stem cells from human exfoliated deciduous teeth differentiate toward neural cells in a medium dynamically cultured with Schwann cells in a series of polydimethylsiloxanes scaffolds. J. Neural Eng. 2016, 13, 046005. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.Y.; Wang, G.S.; Ko, C.S.; Chen, X.W.; Su, W.T. A study of the differentiation of stem cells from human exfoliated deciduous teeth on 3D silk fibroin scaffolds using static and dynamic culture paradigms. Mater. Sci. Eng. C 2020, 109, 110563. [Google Scholar] [CrossRef]
- Dorati, B.; Chiesa, E.; Rosalia, M.; Pisani, S.; Ida Genta, I.; Giovanna Bruni, G.; Modena, T.; Conti, B. Tubular electrospun vancomycin-loaded vascular grafts: Formulation study and physicochemical characterization. Polymers 2021, 13, 2073. [Google Scholar] [CrossRef]
- Huang, T.Y.; Wang, G.S.; Tseng, C.C.; Su, W.T. Epidermal cells differentiated from stem cells from human exfoliated deciduous teeth and seeded onto polyvinyl alcohol/silk fibroin nanofiber dressings accelerate wound repair. Mater. Sci. Eng. C 2019, 104, 109986. [Google Scholar] [CrossRef]
- Li, W.J.; Laurencin, C.T.; Caterson, E.J.; Tuan, R.S.; Ko, F.K. Electrospun nanofibrous structure: A novel scaffold for tissue engineering. J. Biomed. Mater. Res. 2002, 60, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, H.; Shin, Y.M.; Terai, H.; Vacanti, J.P. A biodegradable nanofiber scaffold by electrospinning and its potential for bone tissue engineering. Biomaterials 2003, 4, 2077–2082. [Google Scholar] [CrossRef]
- Alven, S. and Aderibigbe. B.A. Fabrication of Hybrid Nanofibers from Biopolymers and Poly (Vinyl Alcohol)/Poly (ε-Caprolactone) for Wound Dressing Applications. Polymers 2021, 13, 2104. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.S.; Chen, J.H.; Su, W.T. Stem Cells from Human Exfoliated Deciduous Teeth: A Concise Review. Curr. Stem Cell Res. Ther. 2020, 15, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.J.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. Inter. Soc. Cellul. Ther. Posit. State. Cytother. 2006, 8, 315–317. [Google Scholar]
- Seo, B.M.; Miura, M.; Gronthos, S.; Bartold, P.M.; Batouli, S.; Brahim, J.; Young, M.; Robey, P.G.; Wang, C.Y.; Shi, S. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet 2004, 364, 149–155. [Google Scholar] [CrossRef]
- Gay, I.C.; Chen, S.; MacDougall, M. Isolation and characterization of multipotent human periodontal ligament stem cells. Orthod. Craniofac. Res. 2007, 10, 149–160. [Google Scholar] [CrossRef]
- Huang, G.T.; Gronthos, S.; Shi, S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: Their biology and role in regenerative medicine. J. Dent. Res. 2009, 88, 792–806. [Google Scholar] [CrossRef]
- Iwata, T.; Yamato, M.; Zhang, Z.; Mukobata, S.; Washio, K.; Ando, T.; Feijen, J.; Okano, T.; Ishikawa, I. Validation of human periodontal ligament-derived cells as a reliable source for cytotherapeutic use. J. Clin. Periodontol. 2010, 37, 1088–1099. [Google Scholar] [CrossRef] [PubMed]
- Zafar, M.; Najeeb, S.; Khurshid, Z.; Vazirzadeh, M.; Zohaib, S.; Najeeb, B.; Sefat, F. Potential of Electrospun Nanofibers for Biomedical and Dental Applications. Materials 2016, 9, 73. [Google Scholar] [CrossRef] [PubMed]
- Ornitz, D.M.; Marie, P.J. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 2002, 16, 1446–1465. [Google Scholar] [CrossRef] [Green Version]
- Hughes, F.J.; Turner, W.; Belibasakis, G.; Martuscelli, G. Effects of growth factors and cytokines on osteoblast differentiation. Periodontology 2000, 41, 48–72. [Google Scholar] [CrossRef] [PubMed]
- Shin, N.R.; Lee, A.Y.; Park, G.; Ko, J.W.; Kim, J.C.; Shin, I.S.; Kim, J.S. Therapeutic effect of Dipsacus asperoides C. Y. Cheng et T. M. Ai in ovalbumin induced murine model of asthma. Int. J. Mol. Sci. 2019, 20, 1855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, T.M.; Na, M.; Thuong, P.T.; Su, N.D.; Sok, D.; Song, K.S.; Seong, Y.H.; Bae, K. Antioxidant activity of caffeoyl quinic acid derivatives from the roots of Dipsacus asper Wall. J. Ethnopharmacol. 2006, 108, 188–192. [Google Scholar] [CrossRef] [PubMed]
- Moshaverinia, A.; Chen, C.; Xu, X.; Akiyama, K.; Ansari, S.; Zadeh, H.H.; Shi, S. Bone regeneration potential of stem cells derived from periodontal ligament or gingival tissue sources encapsulated in RGD-modified alginate scaffold. Tissue Eng. Part A 2014, 20, 611–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Mu, J.; Fan, Z.; Lei, G.; Yan, M.; Wang, S.; Tang, C.; Wang, Z.; Yu, J.; Zhang, G. Insulin-like growth factor 1 enhances the proliferation and osteogenic differentiation of human periodontal ligament stem cells via ERK and JNK MAPK pathways. Histochem. Cell Biol. 2012, 137, 513–525. [Google Scholar] [CrossRef]
- Ding, G.L.; Liu, Y.; Wang, W.; Wei, F.; Liu, D.; Fan, Z.; An, Y.; Zhang, C.; Wang, S. Allogeneic periodontal ligament stem cell therapy for periodontitis in swine. Stem Cells 2010, 28, 1829–1838. [Google Scholar] [CrossRef] [Green Version]
- Park, J.Y.; Jeon, S.H.; Choung, P.H. Efficacy of periodontal stem cell transplantation in the treatment of advanced periodontitis. Cell Trans. 2011, 20, 271–285. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, J.P.; González, M.; Ríos, S.; Cambiazo, V. Cytoskeletal organization of human mesenchymal stem cells (MSC) changes during their osteogenic differentiation. J. Cell. Biochem. 2004, 93, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Su, W.T.; Chen, P.H. Magnesium and zinc borate enhance osteoblastic differentiation of stem cells from human exfoliated deciduous teeth in vitro. J. Biomater. Appl. 2018, 32, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Deckers, M.M.; van Bezooijen, R.L.; van der Horst, G.; Hoogendam, J.; van Der Bent, C.; Papapoulos, S.E.; Löwik, C.W. Bone morphogenetic proteins stimulate angiogenesis through osteoblast-derived vascular endothelial growth factor A. Endocrinology 2002, 143, 1545–1553. [Google Scholar] [CrossRef] [PubMed]
- Mayr-Wohlfart, U.; Waltenberger, J.; Hausser, H.; Kessler, S.; Günther, K.P.; Dehio, C.; Puhl, W.; Brenner, R.E. Vascular endothelial growth factor stimulates chemotactic migration of primary human osteoblasts. Bone 2002, 30, 472–477. [Google Scholar] [CrossRef]
- Shibuya, M. Vascular endothelial growth factor (VEGF) and its receptor (vegfr) signaling in angiogenesis: A crucial target for anti- and pro-angiogenic therapies. Genes Cancer 2011, 2, 1097–1105. [Google Scholar] [CrossRef]
- Khakestani, M.; Jafari, S.H.; Zahedi, P.; Bagheri, R.; Hajiaghaee, R. Physical, morphological, and biological studies on PLA/nHA composite nanofibrous webs containing Equisetum arvense herbal extract for bone tissue engineering. J. Appl. Polym. Sci. 2017, 134, 45343. [Google Scholar] [CrossRef]
- Pajoumshariati, S.; Yavari, S.K.; Shokrgozar, M.A. Physical and biological modification of polycaprolactone electrospun nanofiber by panax ginseng extract for bone tissue engineering application. Ann. Biomed. Eng. 2016, 44, 1808–1820. [Google Scholar] [CrossRef]
















Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, T.-Y.; Shahrousvand, M.; Hsu, Y.-T.; Su, W.-T. Polycaprolactone/Polyethylene Glycol Blended with Dipsacus asper Wall Extract Nanofibers Promote Osteogenic Differentiation of Periodontal Ligament Stem Cells. Polymers 2021, 13, 2245. https://doi.org/10.3390/polym13142245
Huang T-Y, Shahrousvand M, Hsu Y-T, Su W-T. Polycaprolactone/Polyethylene Glycol Blended with Dipsacus asper Wall Extract Nanofibers Promote Osteogenic Differentiation of Periodontal Ligament Stem Cells. Polymers. 2021; 13(14):2245. https://doi.org/10.3390/polym13142245
Chicago/Turabian StyleHuang, Te-Yang, Mohsen Shahrousvand, Yu-Teng Hsu, and Wen-Ta Su. 2021. "Polycaprolactone/Polyethylene Glycol Blended with Dipsacus asper Wall Extract Nanofibers Promote Osteogenic Differentiation of Periodontal Ligament Stem Cells" Polymers 13, no. 14: 2245. https://doi.org/10.3390/polym13142245