Deciphering the Role of π-Interactions in Polyelectrolyte Complexes Using Rationally Designed Peptides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Polyelectrolyte Complex Preparation
2.3. Nuclear Magnetic Resonance (NMR) Spectroscopy
2.4. Matrix Assisted Laser Desorption/Ionization Time of Flight (MALDI-TOF) Mass Spectrometry
2.5. Circular Dichroism (CD)
2.6. Attenuated Total Reflection-Fourier Transform (ATR-FTIR) Spectroscopy
2.7. Absorbance Measurements
2.8. Optical Microscopy
3. Results and Discussion
3.1. Sequence Patterning of Charged Phenylalanine Peptides
3.2. Circular Dichroism (CD) of Phenylalanine Peptide Sequences
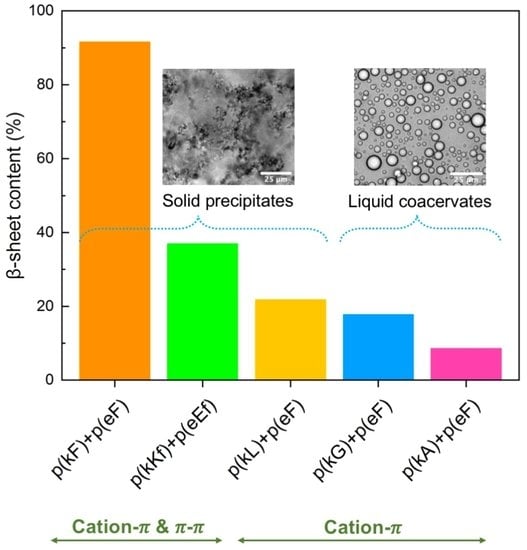
3.3. Secondary Structure Analysis of Polypeptides and Sequence Pairs
3.4. Stability of Sequence Pairs Against Salt
3.5. Compartmentalization of a Small Hydrophobic-Charged Molecule Using Sequence Pairs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Michaels, A.S. Polyelectrolyte complexes. Ind. Eng. Chem. 1965, 57, 32–40. [Google Scholar] [CrossRef]
- Cooper, C.L.; Dubin, P.L.; Kayitmazer, A.B.; Turksen, S. Polyelectrolyte–protein complexes. Curr. Opin. Colloid Interface Sci. 2005, 10, 52–78. [Google Scholar] [CrossRef]
- Thünemann, A.F.; Müller, M.; Dautzenberg, H.; Joanny, J.-F.; Löwen, H. Polyelectrolyte complexes. In Polyelectrolytes with Defined Molecular Architecture II; Schmidt, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2004; pp. 113–171. [Google Scholar]
- Fu, J.; Schlenoff, J.B. Driving forces for oppositely charged polyion association in aqueous solutions: Enthalpic, entropic, but not electrostatic. J. Am. Chem. Soc. 2016, 138, 980–990. [Google Scholar] [CrossRef] [PubMed]
- Priftis, D.; Xia, X.; Margossian, K.O.; Perry, S.L.; Leon, L.; Qin, J.; de Pablo, J.J.; Tirrell, M. Ternary, tunable polyelectrolyte complex fluids driven by complex coacervation. Macromolecules 2014, 47, 3076–3085. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, Y.; Antila, H.S.; Lutkenhaus, J.L.; Sammalkorpi, M. Role of salt and water in the plasticization of pdac/pss polyelectrolyte assemblies. J. Phys. Chem. B 2017, 121, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Van der Gucht, J.; Spruijt, E.; Lemmers, M.; Cohen Stuart, M.A. Polyelectrolyte complexes: Bulk phases and colloidal systems. J. Colloid Interface Sci. 2011, 361, 407–422. [Google Scholar] [CrossRef] [PubMed]
- Mascotti, D.P.; Lohman, T.M. Thermodynamic extent of counterion release upon binding oligolysines to single-stranded nucleic acids. Proc. Natl. Acad. Sci. USA 1990, 87, 3142–3146. [Google Scholar] [CrossRef] [Green Version]
- Sing, C.E.; Perry, S.L. Recent progress in the science of complex coacervation. Soft Matter 2020, 16, 2885–2914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perry, S.L.; Leon, L.; Hoffmann, K.Q.; Kade, M.J.; Priftis, D.; Black, K.A.; Wong, D.; Klein, R.A.; Pierce, C.F., 3rd; Margossian, K.O.; et al. Chirality-selected phase behaviour in ionic polypeptide complexes. Nat. Commun. 2015, 6, 6052. [Google Scholar] [CrossRef]
- Tabandeh, S.; Leon, L. Engineering peptide-based polyelectrolyte complexes with increased hydrophobicity. Molecules 2019, 24, 868. [Google Scholar] [CrossRef] [Green Version]
- Sadman, K.; Wang, Q.; Chen, Y.; Keshavarz, B.; Jiang, Z.; Shull, K.R. Influence of hydrophobicity on polyelectrolyte complexation. Macromolecules 2017, 50, 9417–9426. [Google Scholar] [CrossRef]
- Vieregg, J.R.; Lueckheide, M.; Marciel, A.B.; Leon, L.; Bologna, A.J.; Rivera, J.R.; Tirrell, M.V. Oligonucleotide-peptide complexes: Phase control by hybridization. J. Am. Chem. Soc. 2018, 140, 1632–1638. [Google Scholar] [CrossRef] [PubMed]
- Mende, M.; Schwarz, S.; Zschoche, S.; Petzold, G.; Janke, A. Influence of the hydrophobicity of polyelectrolytes on polyelectrolyte complex formation and complex particle structure and shape. Polymers 2011, 3, 1363–1376. [Google Scholar] [CrossRef] [Green Version]
- Marciel, A.B.; Srivastava, S.; Tirrell, M.V. Structure and rheology of polyelectrolyte complex coacervates. Soft Matter 2018, 14, 2454–2464. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Oh, D.X.; Lee, Y.; Jho, Y.; Hwang, D.S.; Zeng, H. Nanomechanics of cation-pi interactions in aqueous solution. Angew. Chem. Int. Ed. Engl. 2013, 52, 3944–3948. [Google Scholar] [CrossRef]
- Dougherty, D.A. The cation-pi interaction. Acc. Chem. Res. 2013, 46, 885–893. [Google Scholar] [CrossRef] [Green Version]
- Gazit, E. A possible role for pi-stacking in the self-assembly of amyloid fibrils. FASEB J. 2002, 16, 77–83. [Google Scholar] [CrossRef]
- Beene, D.L.; Brandt, G.S.; Zhong, W.; Zacharias, N.M.; Lester, H.A.; Dougherty, D.A. Cation-pi interactions in ligand recognition by serotonergic (5-ht3a) and nicotinic acetylcholine receptors: The anomalous binding properties of nicotine. Biochemistry 2002, 41, 10262–10269. [Google Scholar] [CrossRef]
- Hunter, C.A.; Singh, J.; Thornton, J.M. Π-π interactions: The geometry and energetics of phenylalanine-phenylalanine interactions in proteins. J. Mol. Biol. 1991, 218, 837–846. [Google Scholar] [CrossRef]
- Nott, T.J.; Petsalaki, E.; Farber, P.; Jervis, D.; Fussner, E.; Plochowietz, A.; Craggs, T.D.; Bazett-Jones, D.P.; Pawson, T.; Forman-Kay, J.D.; et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell 2015, 57, 936–947. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Choi, J.M.; Holehouse, A.S.; Lee, H.O.; Zhang, X.; Jahnel, M.; Maharana, S.; Lemaitre, R.; Pozniakovsky, A.; Drechsel, D.; et al. A molecular grammar governing the driving forces for phase separation of prion-like rna binding proteins. Cell 2018, 174, 688–699.e16. [Google Scholar] [CrossRef] [Green Version]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Mahadevi, A.S.; Sastry, G.N. Cation-pi interaction: Its role and relevance in chemistry, biology, and material science. Chem. Rev. 2013, 113, 2100–2138. [Google Scholar] [CrossRef]
- Gomes, E.; Shorter, J. The molecular language of membraneless organelles. J. Biol. Chem. 2019, 294, 7115–7127. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Song, Y.; Sun, W.; Cao, J.; Yuan, H.; Wang, X.; Sun, Y.; Shum, H.C. Cell-inspired all-aqueous microfluidics: From intracellular liquid-liquid phase separation toward advanced biomaterials. Adv. Sci. 2020, 7, 1903359. [Google Scholar] [CrossRef]
- Boeynaems, S.; Alberti, S.; Fawzi, N.L.; Mittag, T.; Polymenidou, M.; Rousseau, F.; Schymkowitz, J.; Shorter, J.; Wolozin, B.; Van Den Bosch, L.; et al. Protein phase separation: A new phase in cell biology. Trends Cell Biol. 2018, 28, 420–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marnik, E.A.; Updike, D.L. Membraneless organelles: P granules in caenorhabditis elegans. Traffic 2019, 20, 373–379. [Google Scholar] [CrossRef] [Green Version]
- Aumiller, W.M., Jr.; Pir Cakmak, F.; Davis, B.W.; Keating, C.D. Rna-based coacervates as a model for membraneless organelles: Formation, properties, and interfacial liposome assembly. Langmuir 2016, 32, 10042–10053. [Google Scholar] [CrossRef] [Green Version]
- Qamar, S.; Wang, G.; Randle, S.J.; Ruggeri, F.S.; Varela, J.A.; Lin, J.Q.; Phillips, E.C.; Miyashita, A.; Williams, D.; Strohl, F.; et al. Fus phase separation is modulated by a molecular chaperone and methylation of arginine cation-pi interactions. Cell 2018, 173, 720–734.e15. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Li, M.; Liu, J. Π–π stacking interaction: A nondestructive and facile means in material engineering for bioapplications. Cryst. Growth Des. 2018, 18, 2765–2783. [Google Scholar] [CrossRef]
- Das, S.; Lin, Y.H.; Vernon, R.M.; Forman-Kay, J.D.; Chan, H.S. Comparative roles of charge, pi, and hydrophobic interactions in sequence-dependent phase separation of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA 2020, 117, 28795–28805. [Google Scholar] [CrossRef]
- Gallivan, J.P.; Dougherty, D.A. Cation-pi interactions in structural biology. Proc. Natl. Acad. Sci. USA 1999, 96, 9459–9464. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, G.; Kartha, K.K.; Fernandez, G. Tuning the mechanistic pathways of peptide self-assembly by aromatic interactions. Chem. Commun. 2021, 57, 1603–1606. [Google Scholar] [CrossRef]
- Wychowaniec, J.K.; Patel, R.; Leach, J.; Mathomes, R.; Chhabria, V.; Patil-Sen, Y.; Hidalgo-Bastida, A.; Forbes, R.T.; Hayes, J.M.; Elsawy, M.A. Aromatic stacking facilitated self-assembly of ultrashort ionic complementary peptide sequence: Beta-sheet nanofibers with remarkable gelation and interfacial properties. Biomacromolecules 2020, 21, 2670–2680. [Google Scholar] [CrossRef]
- Kim, S.; Huang, J.; Lee, Y.; Dutta, S.; Yoo, H.Y.; Jung, Y.M.; Jho, Y.; Zeng, H.; Hwang, D.S. Complexation and coacervation of like-charged polyelectrolytes inspired by mussels. Proc. Natl. Acad. Sci. USA 2016, 113, E847–E853. [Google Scholar] [CrossRef] [Green Version]
- Jayawarna, V.; Ali, M.; Jowitt, T.A.; Miller, A.F.; Saiani, A.; Gough, J.E.; Ulijn, R.V. Nanostructured hydrogels for three-dimensional cell culture through self-assembly of fluorenylmethoxycarbonyl–dipeptides. Adv. Mater. 2006, 18, 611–614. [Google Scholar] [CrossRef]
- Kapelner, R.A.; Yeong, V.; Obermeyer, A.C. Molecular determinants of protein-based coacervates. Curr. Opin. Colloid Interface Sci. 2021, 52, 101407. [Google Scholar] [CrossRef]
- Dougherty, D.A. Cation-pi interactions involving aromatic amino acids. J. Nutr. 2007, 137, 1504S–1508S; discussion 1516S–1517S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, M.; Zacharia, N.S. Sequestration of methylene blue into polyelectrolyte complex coacervates. Macromol. Rapid Commun. 2016, 37, 1249–1255. [Google Scholar] [CrossRef]
- Blocher, W.C.; Perry, S.L. Complex coacervate-based materials for biomedicine. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2017, 9, e1442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Li, C.; Wang, H.Y.; Zhang, X.Z.; Zhuo, R.X. Synthesis of thermo- and ph-sensitive polyion complex micelles for fluorescent imaging. Chemistry 2012, 18, 2297–2304. [Google Scholar] [CrossRef]
- Hoffmann, K.Q.; Perry, S.L.; Leon, L.; Priftis, D.; Tirrell, M.; de Pablo, J.J. A molecular view of the role of chirality in charge-driven polypeptide complexation. Soft Matter 2015, 11, 1525–1538. [Google Scholar] [CrossRef] [PubMed]
- Pacalin, N.M.; Leon, L.; Tirrell, M. Directing the phase behavior of polyelectrolyte complexes using chiral patterned peptides. Eur. Phys. J. Spec. Top. 2016, 225, 1805–1815. [Google Scholar] [CrossRef]
- Lee, C.; Lu, I.C.; Hsu, H.C.; Lin, H.Y.; Liang, S.P.; Lee, Y.T.; Ni, C.K. Formation of metal-related ions in matrix-assisted laser desorption ionization. J. Am. Soc. Mass Spectrom. 2016, 27, 1491–1498. [Google Scholar] [CrossRef]
- Budisa, N.; Wenger, W.; Wiltschi, B. Residue-specific global fluorination of candida antarctica lipase b in pichia pastoris. Mol. Biosyst. 2010, 6, 1630–1639. [Google Scholar] [CrossRef]
- Surewicz, W.K.; Mantsch, H.H.; Chapman, D. Determination of protein secondary structure by fourier transform infrared spectroscopy: A critical assessment. Biochemistry 1993, 32, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krimm, S.; Bandekar, J. Vibrational spectroscopy and conformation of peptides, polypeptides, and proteins. Adv. Protein Chem. 1986, 38, 181–364. [Google Scholar]
- Kong, J.; Yu, S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. 2007, 39, 549–559. [Google Scholar] [CrossRef] [Green Version]
- Ganim, Z.; Chung, H.S.; Smith, A.W.; Deflores, L.P.; Jones, K.C.; Tokmakoff, A. Amide i two-dimensional infrared spectroscopy of proteins. Acc. Chem. Res. 2008, 41, 432–441. [Google Scholar] [CrossRef]
- Coates, J. Interpretation of infrared spectra, a practical approach. In Encyclopedia of Analytical Chemistry; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Haris, P.I.; Chapman, D. The conformational analysis of peptides using fourier transform ir spectroscopy. Biopolymers 1995, 37, 251–263. [Google Scholar] [CrossRef]
- Haris, P.I. Probing protein-protein interaction in biomembranes using fourier transform infrared spectroscopy. Biochim. Biophys. Acta 2013, 1828, 2265–2271. [Google Scholar] [CrossRef]
- Belatik, A.; Hotchandani, S.; Carpentier, R.; Tajmir-Riahi, H.A. Locating the binding sites of pb(ii) ion with human and bovine serum albumins. PLoS ONE 2012, 7, e36723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murthy, A.C.; Dignon, G.L.; Kan, Y.; Zerze, G.H.; Parekh, S.H.; Mittal, J.; Fawzi, N.L. Molecular interactions underlying liquid-liquid phase separation of the fus low-complexity domain. Nat. Struct. Mol. Biol. 2019, 26, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Gabryelczyk, B.; Cai, H.; Shi, X.; Sun, Y.; Swinkels, P.J.M.; Salentinig, S.; Pervushin, K.; Miserez, A. Hydrogen bond guidance and aromatic stacking drive liquid-liquid phase separation of intrinsically disordered histidine-rich peptides. Nat. Commun. 2019, 10, 5465. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Kaplan, D.; Cebe, P. Determining beta-sheet crystallinity in fibrous proteins by thermal analysis and infrared spectroscopy. Macromolecules 2006, 39, 6161–6170. [Google Scholar] [CrossRef]
- Jung, C. Insight into protein structure and protein-ligand recognition by fourier transform infrared spectroscopy. J. Mol. Recognit. 2000, 13, 325–351. [Google Scholar] [CrossRef]
- Gade Malmos, K.; Blancas-Mejia, L.M.; Weber, B.; Buchner, J.; Ramirez-Alvarado, M.; Naiki, H.; Otzen, D. Tht 101: A primer on the use of thioflavin t to investigate amyloid formation. Amyloid 2017, 24, 1–16. [Google Scholar] [CrossRef]
- Biancalana, M.; Koide, S. Molecular mechanism of thioflavin-t binding to amyloid fibrils. Biochim. Biophys. Acta 2010, 1804, 1405–1412. [Google Scholar] [CrossRef] [Green Version]
- Chollakup, R.; Smitthipong, W.; Eisenbach, C.D.; Tirrell, M. Phase behavior and coacervation of aqueous poly(acrylic acid)−poly(allylamine) solutions. Macromolecules 2010, 43, 2518–2528. [Google Scholar] [CrossRef]
- Priftis, D.; Tirrell, M. Phase behaviour and complex coacervation of aqueous polypeptide solutions. Soft Matter 2012, 8, 9396–9405. [Google Scholar] [CrossRef]
- Fu, J.; Fares, H.M.; Schlenoff, J.B. Ion-pairing strength in polyelectrolyte complexes. Macromolecules 2017, 50, 1066–1074. [Google Scholar] [CrossRef]
- Chollakup, R.; Beck, J.B.; Dirnberger, K.; Tirrell, M.; Eisenbach, C.D. Polyelectrolyte molecular weight and salt effects on the phase behavior and coacervation of aqueous solutions of poly(acrylic acid) sodium salt and poly(allylamine) hydrochloride. Macromolecules 2013, 46, 2376–2390. [Google Scholar] [CrossRef]
- Dautzenberg, H.; Karibyants, N. Polyelectrolyte complex formation in highly aggregating systems. Effect of salt: Response to subsequent addition of nacl. Macromol. Chem. Phys. 1999, 200, 118–125. [Google Scholar] [CrossRef]
- Perry, S.; Li, Y.; Priftis, D.; Leon, L.; Tirrell, M. The effect of salt on the complex coacervation of vinyl polyelectrolytes. Polymers 2014, 6, 1756–1772. [Google Scholar] [CrossRef] [Green Version]
- Kayitmazer, A.B.; Koksal, A.F.; Kilic Iyilik, E. Complex coacervation of hyaluronic acid and chitosan: Effects of ph, ionic strength, charge density, chain length and the charge ratio. Soft Matter 2015, 11, 8605–8612. [Google Scholar] [CrossRef]
- Qin, J.; Priftis, D.; Farina, R.; Perry, S.L.; Leon, L.; Whitmer, J.; Hoffmann, K.; Tirrell, M.; de Pablo, J.J. Interfacial tension of polyelectrolyte complex coacervate phases. ACS Macro Lett. 2014, 3, 565–568. [Google Scholar] [CrossRef]
- Li, L.; Srivastava, S.; Meng, S.; Ting, J.M.; Tirrell, M.V. Effects of non-electrostatic intermolecular interactions on the phase behavior of ph-sensitive polyelectrolyte complexes. Macromolecules 2020, 53, 7835–7844. [Google Scholar] [CrossRef]
- Wang, Q.; Schlenoff, J.B. The polyelectrolyte complex/coacervate continuum. Macromolecules 2014, 47, 3108–3116. [Google Scholar] [CrossRef]
- Kang, B.; Tang, H.; Zhao, Z.; Song, S. Hofmeister series: Insights of ion specificity from amphiphilic assembly and interface property. ACS Omega 2020, 5, 6229–6239. [Google Scholar] [CrossRef] [Green Version]
- Baldwin, R.L. How hofmeister ion interactions affect protein stability. Biophys. J. 1996, 71, 2056–2063. [Google Scholar] [CrossRef] [Green Version]
- Krainer, G.; Welsh, T.J.; Joseph, J.A.; Espinosa, J.R.; Wittmann, S.; de Csillery, E.; Sridhar, A.; Toprakcioglu, Z.; Gudiskyte, G.; Czekalska, M.A.; et al. Reentrant liquid condensate phase of proteins is stabilized by hydrophobic and non-ionic interactions. Nat. Commun. 2021, 12, 1085. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Rumyantsev, A.M.; Srivastava, S.; Meng, S.; de Pablo, J.J.; Tirrell, M.V. Effect of solvent quality on the phase behavior of polyelectrolyte complexes. Macromolecules 2020, 54, 105–114. [Google Scholar] [CrossRef]
- Pak, C.W.; Kosno, M.; Holehouse, A.S.; Padrick, S.B.; Mittal, A.; Ali, R.; Yunus, A.A.; Liu, D.R.; Pappu, R.V.; Rosen, M.K. Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell 2016, 63, 72–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]









| Polycations | m/z (g/mol) | Polyanions | m/z (g/mol) |
|---|---|---|---|
| (kF)15 | 4174.24 | (eF)15 | 4185.57 |
| (kKfKkF)5 | 4077.47 | (eEfEeF)5 | 4097.66 |
| (k(fl)F)15 | 4443.6 | (e(fl)F)15 | 4460.08 |
| Secondary Structure 1 | Sequence Pairs | ||
|---|---|---|---|
| p(kF) + p(eF) | p(kKf) + p(eEf) | p(k(fl)F) + p(e(fl)F) | |
| β-Sheet (1615–1640 cm−1) | 73.2% | 31.5% | 24% |
| Random Coil (1639–1654 cm−1) | 8.4% | 63% | 76% |
| β-Sheet (1670–1694 cm−1) | 18.4% 2 | 5.5% | - |
| Secondary Structure 1 | Complex Pairs | |||
|---|---|---|---|---|
| p(kG) + p(eF) | p(kA) + p(eF) | p(kL) + p(eF) | p(kL) + p(e(fl)F) | |
| β-Sheet (1615–1640 cm−1) | - | - | 21.77% 2 | 30.1% |
| Random Coil (1639–1654 cm−1) | 82.2% | 91.4% | 78.23% | 65.5% |
| β-Sheet (1670–1694 cm−1) | 17.8% | 8.6% | - | 4.4% |
| Sequence Pairs | EE (%) | Sequence Pairs | EE (%) |
|---|---|---|---|
| p(kF) + p(eF) | 42.7 | p(kG) + p(eF) | 18.05 |
| p(kKf) + p(eEf) | 99.15 | p(kA) + p(eF) | 21.25 |
| p(k(fl)F) + p(e(fl)F) | 27.34 | p(kL) + p(eF) | 28.83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tabandeh, S.; Lemus, C.E.; Leon, L. Deciphering the Role of π-Interactions in Polyelectrolyte Complexes Using Rationally Designed Peptides. Polymers 2021, 13, 2074. https://doi.org/10.3390/polym13132074
Tabandeh S, Lemus CE, Leon L. Deciphering the Role of π-Interactions in Polyelectrolyte Complexes Using Rationally Designed Peptides. Polymers. 2021; 13(13):2074. https://doi.org/10.3390/polym13132074
Chicago/Turabian StyleTabandeh, Sara, Cristina Elisabeth Lemus, and Lorraine Leon. 2021. "Deciphering the Role of π-Interactions in Polyelectrolyte Complexes Using Rationally Designed Peptides" Polymers 13, no. 13: 2074. https://doi.org/10.3390/polym13132074