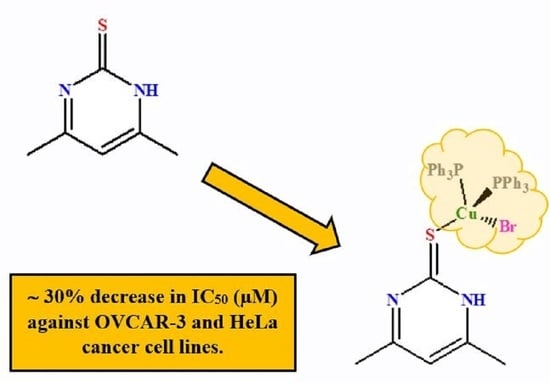
Synthesis, Structural Studies, and Anticancer Properties of [CuBr(PPh3)2(4,6-Dimethyl-2-Thiopyrimidine-κS]
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Instrumentation
2.3. Synthesis and Characterization
2.4. Single-Crystal X-ray Diffraction Study
2.5. HSA Binding Studies
2.6. Molecular Docking
2.7. Anticancer Studies
3. Results
3.1. Synthesis and Characterization
3.2. Crystal Structure Study
3.3. Protein Binding Study
3.4. Molecular Docking
3.5. Anticancer Activities
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
References
- Raper, E.S. Complexes of heterocyclic thionates Part 2: Complexes of bridging ligands. Coord. Chem. Rev. 1997, 165, 475–567. [Google Scholar] [CrossRef]
- Garcia-Vazquez, J.A.; Romero, J.; Sousa, A. Electrochemical synthesis of metallic complexes of bidentate thiolates containing nitrogen as an additional donor atom. Coord. Chem. Rev. 1999, 193, 691–745. [Google Scholar] [CrossRef]
- Akrivos, P.D. Recent studies in the coordination chemistry of heterocyclic thiones and thionates. Coord. Chem. Rev. 2001, 213, 181–210. [Google Scholar] [CrossRef]
- Karagiannidis, P.; Aslanidis, P.; Kessissoglou, D.P.; Krebs, B.; Dartmann, M. Mononuclear and binuclear Cu(I) complexes with metal-sulfur ligation. Inorg. Chim. Acta 1989, 156, 47–56. [Google Scholar] [CrossRef]
- Skoulika, S.; Aubry, A.; Karagianidis, P.; Aslanidis, P.; Papastefanou, S. New copper(I) chloride complexes with heterocyclic thiones and triphenylphosphine as ligands. Crystal structures of [Cu(PPh3)2(bzimtH2)Cl·CH3COCH3 and [Cu(PPh3)2(nbzimtH2)Cl]. Inorg. Chim. Acta 1991, 183, 207–211. [Google Scholar] [CrossRef]
- Batsala, G.K.; Dokorou, V.; Kourkoumelis, N.; Manos, M.J.; Tasiopoulos, A.J.; Mavromoustakos, T.; Simčic, M.; Golič-Grdadolnik, S.; Hadjikakou, S.K. Copper(I)/(II) or silver(I) ions towards 2-mercaptopyrimidine: An exploration of a chemical variability with possible biological implication. Inorg. Chim. Acta 2012, 382, 146–157. [Google Scholar] [CrossRef]
- Anastasiadou, D.; Psomas, G.; Lalia-Kantouri, M.; Hatzidimitriou, A.G.; Aslanidis, P. Copper(I) halide complexes of 2,2,5,5-tetramethyl-imidazolidine-4-thione: Synthesis, structures, luminescence, thermal stability and interaction with DNA. Mater. Sci. Eng. C 2016, 68, 241–250. [Google Scholar] [CrossRef]
- Hadjikakou, S.K.; Aslanidis, P.; Karagiannidis, P.; Mentzafos, D.; Terzis, A. Synthesis and photolysis of mixed copper(I) complexes with thiones and tri-p-tolylphosphine or triphenylphosphine; X-ray crystal structure of bis[copper(I)(1,3-thiazolidine-2-thione) (tri-p-tolylphosphine)chloride]. Polyhedron 1991, 10, 935–940. [Google Scholar] [CrossRef]
- Hadjikakou, S.K.; Aslanidis, P.; Akrivos, P.D.; Karagiannidis, P.; Kojic-Prodic, B.; Luic, M. Study of mixed ligand copper(I) complexes with tri-m-tolyl-phosphine (tmtp) and heterocyclic thiones. Crystal structures of bis[μ-S(benzimidazoline-2-thione)(tmtp) copper(I) chloride] and bis[μ-Br(thiazolidine-2-thione)(tmtp) copper(I)]. Inorg. Chim. Acta 1992, 197, 31–38. [Google Scholar] [CrossRef]
- Hadjikakou, S.K.; Aslanidis, P.; Karagiannidis, P.; Aubry, A.; Skoulika, S. Copper(I) complexes with tri-o-tolylphosphine and heterocyclic thione ligands. Crystal structures of [(pyrimidine-2-thione)(tri-o-tolylphosphine)copper(I) chloride] and [(pyridine-2-thione)(tri-o-tolylphosphine)copper(I) iodide]. Inorg. Chim. Acta 1992, 193, 129–135. [Google Scholar] [CrossRef]
- Walia, S.; Kaur, S.; Kaur, J.; Sandhu, A.K.; Lobana, T.S.; Hundal, G.; Jasinski, J.P. Synthesis, Spectroscopy, and Structures of Mono- and Dinuclear Copper(I) Halide Complexes with 1,3-Imidazolidine-2-thiones. Z. Anorg. Allg. Chem. 2015, 641, 1728–1736. [Google Scholar] [CrossRef]
- Aslanidis, P.; Cox, P.J.; Divanidis, S.; Tsipis, A.C. Copper(I) Halide Complexes with 1,3-Propanebis(diphenylphosphine) and Heterocyclic Thione Ligands: Crystal and Electronic Structures (DFT) of [CuCl(pymtH)(dppp)], [CuBr(pymtH)(dppp)], and [Cu(μ-I)(dppp)]2. Inorg. Chem. 2002, 41, 6875–6886. [Google Scholar] [CrossRef] [PubMed]
- Aslanidis, P.; Cox, P.J.; Divanidis, S.; Karagiannidis, P. Copper(I) halide complexes from cis-1,2-bis(diphenylphosphino) ethylene and some heterocyclic thiones. Inorg. Chim. Acta 2004, 357, 1063–1076. [Google Scholar] [CrossRef]
- Cox, P.J.; Kaltzoglou, A.; Aslanidis, P. Copper(I) halide chelates of the wide bite angle diphosphane xantphos: Crystal structures of [CuBr(xantphos)(dmpymtH)] and [CuI(xantphos)(imdtH2)].CH3CN. Inorg. Chim. Acta 2006, 359, 3183–3190. [Google Scholar] [CrossRef]
- Aslanidis, P.; Cox, P.J.; Tsaliki, P. Copper(I) halide complexes with 2,2′-bis(diphenylphosphano)-1,1′-binaphthyl (rac-binap) and heterocyclic thiones. Racemic compounds in chiral and achiral crystal space groups. Polyhedron 2008, 27, 3029–3035. [Google Scholar] [CrossRef]
- Rodríguez, A.; Sousa-Pedrares, A.; García-Vázquez, J.A.; Romero, J.; Sousa, A. Synthesis and Structural Characterization of Copper(I), Silver(I) and Gold(I) Complexes with Pyrimidine-2-thionato Ligands and their Adducts with Phosphanes. Eur. J. Inorg. Chem. 2011, 2011, 3403–3413. [Google Scholar] [CrossRef]
- Tisato, F.; Porchia, M.; Santini, C.; Gandin, V.; Marzano, C. Ch 3—Phosphine–copper(I) complexes as anticancer agents: Design, synthesis, and physicochemical characterization. Part I. In Copper(I) Chemistry of Phosphines, Functionalized Phosphines and Phosphorus Heterocycles; Balakrishna, M.S., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 61–82. [Google Scholar]
- Marzano, C.; Gandin, V.; Pellei, M.; Colavito, D.; Papini, G.; Lobbia, G.G.; Giudice, E.D.; Porchia, M.; Tisato, F.; Santini, C. In vitro antitumor activity of the water soluble copper (I) complexes bearing the tris (hydroxymethyl) phosphine ligand. J. Med. Chem. 2008, 51, 798–808. [Google Scholar] [CrossRef]
- Lazarou, K.; Bednarz, B.; Kubicki, M.; Verginadis, I.I.; Charalabopoulos, K.; Kourkoumelis, N.; Hadjikakou, S.K. Structural, photolysis and biological studies of the bis (μ2-chloro)-tris (triphenylphosphine)-di-copper (I) and chloro-tris (triphenylphosphine)-copper (I) complexes. Study of copper (I)–copper (I) interactions. Inorg. Chim. Acta 2010, 363, 763–772. [Google Scholar] [CrossRef]
- Porchia, M.; Dolmella, A.; Gandin, V.; Marzano, C.; Pellei, M.; Peruzzo, V.; Refosco, F.; Santini, C.; Tisato, F. Neutral and charged phosphine/scorpionate copper (I) complexes: Effects of ligand assembly on their antiproliferative activity. Eur. J. Med. Chem. 2013, 59, 218–226. [Google Scholar] [CrossRef]
- Gandin, V.; Tisatom, F.; Dolmella, A.; Pellei, M.; Santini, C.; Giorgetti, M.; Marzano, C.; Porchia, M. In vitro and in vivo anticancer activity of copper (I) complexes with homoscorpionate tridentate tris (pyrazolyl) borate and auxiliary monodentate phosphine ligands. J. Med. Chem. 2014, 57, 4745–4760. [Google Scholar] [CrossRef]
- Komarnicka, U.K.; Starosta, R.; Kyziol, A.; Jezowska-Bojczuk, M. Copper (I) complexes with phosphine derived from sparfloxacin. Part I–structures, spectroscopic properties and cytotoxicity. Dalton Trans. 2015, 44, 12688–12699. [Google Scholar] [CrossRef]
- Komarnicka, U.K.; Starosta, R.; Plotek, M.; de Almeida, R.F.M.; Jezowska-Bojczuk, M.; Kyziol, A. Copper (I) complexes with phosphine derived from sparfloxacin. Part II: A first insight into the cytotoxic action mode. Dalton Trans. 2016, 45, 5052–5063. [Google Scholar] [CrossRef]
- Komarnicka, U.K.; Starosta, R.; Kyziol, A.; Plotek, M.; Puchalska, M.; Jezowska-Bojczuk, M. New copper (I) complexes bearing lomefloxacin motif: Spectroscopic properties, in vitro cytotoxicity and interactions with DNA and human serum albumin. J. Inorg. Biochem. 2016, 165, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Mashat, K.; Babgi, B.A.; Hussien, M.A.; Arshad, M.N.; Abdellattif, M. Synthesis, structures, DNA-binding and anticancer activities of some copper(I)-phosphine complexes. Polyhedron 2019, 158, 164–172. [Google Scholar] [CrossRef]
- Babgi, B.A.; Mashat, K.H.; Abdellattif, M.H.; Arshad, M.N.; Alzahrani, K.A.; Asiri, A.M.; Du, J.; Humphrey, M.G.; Hussien, M.A. Synthesis, structures, DNA-binding, cytotoxicity and molecular docking of CuBr (PPh3)(diimine). Polyhedron 2020, 192, 114847. [Google Scholar] [CrossRef]
- Alsaedi, S.; Babgi, B.A.; Abdellatif, M.H.; Arshad, M.N.; Emwas, A.; Jaremko, M.; Humphrey, M.G.; Asiri, A.M.; Hussein, M.A. DNA-Binding and Cytotoxicity of Copper(I) Complexes Con-taining Functionalized Dipyridylphenazine Ligands. Pharmaceutics 2021, 13, 764-1–764-13. [Google Scholar] [CrossRef] [PubMed]
- Aslanidis, P.; Cox, P.J.; Kapetangiannis, K.; Tsipis, A.C. Structural and Spectroscopic Properties of New Copper(I) Complexes with 1,1,1-Tris(diphenylphosphanylmethyl)ethane and Heterocyclic Thiolates. Eur. J. Inorg. Chem. 2008, 2008, 5029–5037. [Google Scholar] [CrossRef]
- Khan, A.; Jasinski, J.P.; Smoleaski, V.A.; Paul, K.; Singh, G.; Sharma, R. Synthesis, structure and cytotoxicity evaluation of complexes of N1-substituted-isatin-3-thiosemicarbazone with copper(I) halides. Inorg. Chim. Acta 2016, 449, 119–126. [Google Scholar] [CrossRef] [Green Version]
- Papazoglou, I.; Cox, P.J.; Hatzidimitriou, A.G.; Kokotidou, C.; Choli-Papadopoulou, T.; Aslanidis, P. Copper(I) halide complexes of 5-carbethoxy-2-thiouracil: Synthesis, structure and in vitro cytotoxicity. Eur. J. Med. Chem. 2014, 78, 383–391. [Google Scholar] [CrossRef]
- Sahu, M.; Siddiqui, N. A review on biological importance of pyrimidines in the new era. Int. J. Pharm. Pharm. Sci. 2016, 8, 8–21. [Google Scholar]
- Martin, D.S.; Bertino, J.R.; Koutcher, J.A. ATP Depletion + Pyrimidine Depletion Can Markedly Enhance Cancer Therapy: Fresh Insight for a New Approach. Cancer Res. 2000, 60, 6776–6783. [Google Scholar]
- Fargualy, A.M.; Habib, N.S.; Ismail, K.A.; Hassan, A.M.M.; Sarg, M.T.M. Synthesis, Biological Evaluation and Molecular Docking Studies of Some Pyrimidine Derivatives. Eur. J. Med. Chem. 2013, 66, 276–295. [Google Scholar] [CrossRef] [PubMed]
- Helwa, A.A.; Gedawy, E.M.; Abou-Seri, S.M.; Taher, A.T.; El-Ansary, A.K. Synthesis and bioactivity evaluation of new pyrimidinone-5-carbonitriles as potential anticancer and antimicrobial agents. Res. Chem. Intermed. 2018, 44, 2685–2702. [Google Scholar] [CrossRef]
- Al-Masri, H.T.; Emwas, A.M.; Al-Talla, Z.A.; Alkordi, M.H. Synthesis and Characterization of New N-(Diphenylphosphino)-Naphthylamine Chalcogenides: X-Ray Structures of (1-NHC10H7)P(Se)Ph2 and Ph2P(S)OP(S)Ph2. Phosphorus Sulfur Silicon Relat. Elem. 2012, 187, 1082–1090. [Google Scholar] [CrossRef]
- Sheldrick, G. SHELXT—Integrated space-group and crystal-structure determination. Acta Cryst. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G. Crystal structure refinement with SHELXL. Acta Cryst. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Cryst. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Hoefelschweiger, B.K.; Duerkop, A.; Wolfbeis, O.S. Novel type of general protein assay using a chromogenic and fluorogenic amine-reactive probe. Anal. Biochem. 2005, 344, 122–129. [Google Scholar] [CrossRef]
- Petitpas, I.; Bhattacharya, A.A.; Twine, S.; East, M.; Curryi, S. Crystal structure analysis of warfarin binding to human serum albumin: Anatomy of drug site I. J. Biol. Chem. 2001, 276, 22804–22809. [Google Scholar] [CrossRef] [Green Version]
- Al-Khathami, N.D.; Al-Rashdi, K.S.; Babgi, B.A.; Hussien, M.A.; Arshad, M.N.; Eltayeb, N.E.; Elsilk, S.E.; Lasri, J.; Basaleh, A.S.; Al-Jahdali, M. Spectroscopic and biological properties of platinum complexes derived from 2-pyridyl Schiff bases. J. Saudi Chem. Soc. 2019, 23, 903–915. [Google Scholar] [CrossRef]
- Abdel-Rhman, M.H.; Hussien, M.A.; Mahmoud, H.M.; Hosny, N.M. Synthesis, characterization, molecular docking and cytotoxicity studies on N-benzyl-2-isonicotinoylhydrazine-1-carbothioamide and its metal complexes. J. Mol. Struct. 2019, 1196, 417–428. [Google Scholar] [CrossRef]
- Cole, J.C.; Murray, C.W.; Nissink, J.W.; Taylor, R.D.; Taylor, R. Comparing protein-ligand docking programs is difficult. Proteins 2005, 60, 325–332. [Google Scholar] [CrossRef]
- Jain, A.N. Bias, reporting, and sharing: Computational evaluations of docking methods. J. Comput. Aided Mol. Des. 2008, 22, 201–212. [Google Scholar] [CrossRef]
- Muanza, D.N.; Kim, B.W.; Euler, K.L.; Williams, L. Antibacterial and Antifungal Activities of Nine Medicinal Plants from Zaire. Int. J. Pharm. 1994, 32, 337–345. [Google Scholar] [CrossRef]
- Pezzuto, J.M.; Che, C.-T.; McPherson, D.D.; Zhu, J.-P.; Topcu, G.; Erdelmeier, C.A.J.; Cordell, G.A. DNA as an Affinity Probe Useful in the Detection and Isolation of Biologically Active Natural Products. J. Nat. Prod. 1991, 54, 1522–1530. [Google Scholar] [CrossRef] [PubMed]
- Skehan, P.; Storeng, R.; Scudiero, D.; Monks, A.; McMahon, J.; Vistica, D.; Warren, J.; Bokesch, H.; Kenney, S.; Boyd, M. New Colorimetric Cytotoxicity Assay for Anticancer-Drug Screening. J. Natl. Cancer Inst. 1990, 82, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Lobana, T.S.; Sandhu, A.K.; Sultana, R.; Castineiras, A.; Butcher, R.J.; Jasinski, J.P. Coordination variability of Cuᴵ in multidonor heterocyclic thioamides: Synthesis, crystal structures, luminescent properties and ESI-mass studies of complexes. RSC Adv. 2014, 4, 30511–30522. [Google Scholar] [CrossRef]
- Anastasiadou, D.; Psomas, G.; Kalogiannis, S.; Geromichalos, G.; Hatzidimitriou, A.G.; Aslanidis, P. Bi- and trinuclear copper(I) compounds of 2,2,5,5-tetramethylimidazolidine-4-thione and 1,2-bis(diphenylphosphano)ethane: Synthesis, crystal structures, in vitro and in silico study of antibacterial activity and interaction with DNA and albumins. J. Inorg. Biochem. 2019, 110750. [Google Scholar] [CrossRef] [PubMed]
- Charalampou, D.C.; Kourkoumelis, N.; Karanestora, S.; Hadjiarapoglou, L.P.; Dokorou, V.; Skoulika, S.; Owczarzak, A.; Kubicki, M.; Hadjikakou, S.K. Mono- and Binuclear Copper(I) Complexes of Thionucleotide Analogues and Their Catalytic Activity on the Synthesis of Dihydrofurans. Inorg. Chem. 2014, 53, 8322–8333. [Google Scholar] [CrossRef] [PubMed]
- Koutsari, A.; Karasmani, F.; Kapetanaki, E.; Zainuddin, D.I.; Hatzidimitriou, A.G.; Angaridis, P.; Aslanidis, P. Luminescent thione/phosphane mixed-ligand copper(I) complexes: The effect of thione on structural properties. Inorg. Chim. Acta 2017, 458, 138–145. [Google Scholar] [CrossRef]
- Lecomte, C.; Skoulika, S.; Aslanidis, P.; Karagiannidis, P.; Papastefanou, S. Copper(I) bromide complexes with heterocyclic thiones and triphenylphosphine as ligands. The X-ray crystal structure of copper(I) pyrimidine-2-thione bis (triphenylphosphine) bromide [Cu(PPh3)2(PymtH)Br]. Polyhedron 1989, 8, 1103–1109. [Google Scholar] [CrossRef]
- Ascone, I.; Messori, L.; Casini, A.; Gabbiani, C.; Balerna, A.; Dell’Unto, F.; Castellano, A.C. Exploiting Soft and Hard X-Ray Absorption Spectroscopy to Characterize Metallodrug/Protein Interactions: The Binding of [trans-RuCl4(Im)(dimethylsulfoxide)][ImH] (Im = imidazole) to Bovine Serum Albumin. Inorg. Chem. 2008, 47, 8629–8634. [Google Scholar] [CrossRef] [PubMed]
- Groessl, M.; Terenghi, M.; Casini, A.; Elviri, L.; Lobinski, R.; Dyson, P.J. Reactivity of anticancer metallodrugs with serum proteins: New insights from size exclusion chromatography-ICP-MS and ESI-MS. J. Anal. At. Spectrom. 2010, 25, 305–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shmykov, A.Y.; Filippov, V.N.; Foteeva, L.S.; Keppler, B.K.; Timerbaev, A.R. Toward high-throughput monitoring of metallodrug–protein interaction using capillary electrophoresis in chemically modified capillaries. Anal. Biochem. 2008, 379, 216–218. [Google Scholar] [CrossRef] [PubMed]
- Timerbaev, A.R.; Hartinger, C.G.; Aleksenko, S.S.; Keppler, B.K. Interactions of Antitumor Metallodrugs with Serum Proteins: Advances in Characterization Using Modern Analytical Methodology. Chem. Rev. 2006, 106, 2224–2248. [Google Scholar] [CrossRef] [PubMed]
- Gibellini, D.; Vitone, F.; Schiavone, P.; Ponti, C.; Placa, M.L.; Re, M.C. Quantitative detection of human immunodeficiency virus type 1 (HIV-1) proviral DNA in peripheral blood mononuclear cells by SYBR green real-time PCR technique. J. Clin. Virol. 2004, 29, 282. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, NY, USA, 2006. [Google Scholar]
- Al-Harthi, S.; Lachowicz, J.I.; Nowakowski, M.E.; Jaremko, M.; Jaremko, Ł. Towards the functional high-resolution coordination chemistry of blood plasma human serum albumin. J. Inorg. Biochem. 2019, 198, 110716. [Google Scholar] [CrossRef]
- Helms, M.K.; Petersen, C.E.; Bhagavan, N.V.; Jameson, D.M. Time-resolved fluorescence studies on site-directed mutants of human serum albumin. FEBS Lett. 1997, 408, 67–70. [Google Scholar] [CrossRef] [Green Version]
- El-Kemary, M.; Gil, M.; Douhal, A. Relaxation Dynamics of Piroxicam Structures within Human Serum Albumin Protein. J. Med. Chem. 2007, 50, 2896–2902. [Google Scholar] [CrossRef]
- He, X.M.; Carter, D.C. Atomic structure and chemistry of human serum albumin. Nature 1992, 358, 209–215. [Google Scholar] [CrossRef] [Green Version]
- Lhiaubet-Vallet, V.; Sarabia, Z.; Boscá, F.; Miranda, M.A. Human Serum Albumin-Mediated Stereodifferentiation in the Triplet State Behavior of (S)- and (R)-Carprofen. J. Am. Chem. Soc. 2004, 126, 9538–9539. [Google Scholar] [CrossRef] [PubMed]
- Yeggoni, D.P.; Gokara, M.; Manidhar, D.M.; Rachamallu, A.; Nakka, S.; Reddy, C.S.; Subramanyam, R. Binding and Molecular Dynamics Studies of 7-Hydroxycoumarin Derivatives with Human Serum Albumin and Its Pharmacological Importance. Mol. Pharm. 2014, 11, 1117–1131. [Google Scholar] [CrossRef] [PubMed]
- Shahabadi, N.; Kashanian, S.; Shalmashi, K.; Roshanfekr, H. DNA Interaction with PtCl2(LL) (LL = Chelating Diamine Ligand: N,N-Dimethyltrimethylendiamine) Complex. Appl. Biochem. Biotechnol. 2009, 158, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Lim-Wilby, M. Molecular docking. In Molecular Modeling of Proteins; Humana Press: Totowa, NJ, USA, 2008; Volume 443, pp. 365–382. [Google Scholar]
- Bhattacharya, B.; Nakka, S.; Guruprasad, L.; Samanta, A. Interaction of Bovine Serum Albumin with Dipolar Molecules: Fluorescence and Molecular Docking Studies. J. Phys. Chem. B 2009, 113, 2143–2150. [Google Scholar] [CrossRef] [PubMed]
- Guizado, T.R.C.; Louro, S.R.W.; Anteneodo, C. Dynamics of heme complexed with human serum albumin: A theoretical approach. Eur. Biophys. J. 2012, 41, 1033–1042. [Google Scholar] [CrossRef]
- Badisa, R.B.; Darling-Reed, S.F.; Joseph, P.; Cooperwood, J.S.; Latinwo, L.M.; Goodman, C.B. Selective Cytotoxic Activities of Two Novel Synthetic Drugs on Human Breast Carcinoma MCF-7 Cells. Anticancer Res. 2009, 29, 2993–2996. [Google Scholar]




| Empirical formula | C42H38BrCuN2P2S | ||
| Formula weight | 808.19 | ||
| Temperature/K | 120 | ||
| Crystal system | Monoclinic | ||
| Space group | P21/c | ||
| a/Å | 9.7089(4) | ||
| b/Å | 17.8054(6) | ||
| c/Å | 21.9786(8) | ||
| β/° | 101.040(2) | ||
| Volume/Å3 | 3729.1(2) | ||
| Z | 4 | ||
| ρcalc g/cm3 | 1.440 | ||
| μ/mm−1 | 1.832 | ||
| F(000) | 1656.0 | ||
| Crystal size/mm3 | 0.355 × 0.113 × 0.11 | ||
| Radiation/Å | MoKα (λ = 0.71073) | ||
| Absorption correction | numerical | ||
| Index ranges | −16 ≤ h ≤ 17/−31 ≤ k ≤ 31/−39 ≤ l ≤ 39 | ||
| Reflections collected | 126915 | ||
| Rint/Rsigma | 0.0506/0.0426 | ||
| Data/restraints/parameters | 22262/214/532 | ||
| Goodness-of-fit on F2 | 1.015 | ||
| R1/wR2 | 0.0358/0.0775 | ||
| Largest diff. peak/hole/e Å−3 | 0.81/−1.00 |
| Compound | Binding Domain | Binding Score | Donor → Acceptor |
|---|---|---|---|
| R-warfarin | II A | −6.27 | (OH)aliph → His288 |
| Arg257 → (OH)aliph | |||
| L | I B/II A | −4.83 | (NH) → Cys200 |
| Lys199 → (S) | |||
| Gln196 → (S) | |||
| Cu-L | II A | −8.38 | Lys199 → (S) |
| Arg222 → (Br) |
| Compound | OVCAR-3 (IC50% in µM) | HeLa (IC50% in µM) | LLC-MK2 (IC50% in µM) |
|---|---|---|---|
| L | 21.35 ± 1.30 | 19.37 ± 0.71 | >100 |
| Cu-L | 15.12 ± 0.48 | 13.05 ± 0.61 | >100 |
| cisplatin | 11.87 ± 1.91 | 11.45 ± 0.60 | >100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Babgi, B.A.; Alsayari, J.H.; Davaasuren, B.; Emwas, A.-H.; Jaremko, M.; Abdellattif, M.H.; Hussien, M.A. Synthesis, Structural Studies, and Anticancer Properties of [CuBr(PPh3)2(4,6-Dimethyl-2-Thiopyrimidine-κS]. Crystals 2021, 11, 688. https://doi.org/10.3390/cryst11060688
Babgi BA, Alsayari JH, Davaasuren B, Emwas A-H, Jaremko M, Abdellattif MH, Hussien MA. Synthesis, Structural Studies, and Anticancer Properties of [CuBr(PPh3)2(4,6-Dimethyl-2-Thiopyrimidine-κS]. Crystals. 2021; 11(6):688. https://doi.org/10.3390/cryst11060688
Chicago/Turabian StyleBabgi, Bandar A., Jalal H. Alsayari, Bambar Davaasuren, Abdul-Hamid Emwas, Mariusz Jaremko, Magda H. Abdellattif, and Mostafa A. Hussien. 2021. "Synthesis, Structural Studies, and Anticancer Properties of [CuBr(PPh3)2(4,6-Dimethyl-2-Thiopyrimidine-κS]" Crystals 11, no. 6: 688. https://doi.org/10.3390/cryst11060688