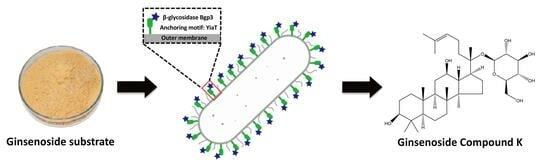
The Synthesis of Ginsenoside Compound K Using a Surface-Displayed β-Glycosidase Whole-Cell Catalyst
Abstract
:1. Introduction
2. Results and Discussion
2.1. Expression and Analysis of Recombinant Proteins in E. coli
2.2. Confirmation of the Surface Display System
2.3. Determination of Surface Display Efficiency
2.4. Effects of IPTG Concentration on Protein Expression and Ginsenoside CK Production
2.5. Effects of Induction Temperature on Protein Expression and Ginsenoside CK Production
2.6. Effects of Ginsenoside Substrate Concentration on Ginsenoside CK Production
2.7. Effects of Catalytic Temperature on Ginsenoside CK Production
2.8. Synthesis of Ginsenoside CK in a 100 mL System
2.9. Evaluation of the Reusability of the Surface Display System
3. Materials and Methods
3.1. Materials
3.2. Strains and Plasmids
3.3. Plasmid Construction and Transformation
3.4. Culture Conditions
3.5. Ginsenoside CK Synthesis
3.6. Analytical Methods
3.6.1. Biomass Determination
3.6.2. High-Performance Liquid Chromatography (HPLC) Analysis of Ginsenoside CK
3.6.3. Trypsin Accessibility Assay for Confirmation of the Surface Display System
3.6.4. Enzymatic Activity Assays
3.6.5. Data Analysis
3.7. Effects of IPTG Concentration and Induction Temperature on Protein Expression and Ginsenoside CK Production
3.8. Effects of Ginsenoside Substrate Concentration and Catalytic Temperature on Ginsenoside CK Production
3.9. Scale-Up of Ginsenoside CK Synthesis Reaction
3.10. Reusability Assay of Surface Display System
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, P.; Tang, C.; Liu, Y.; Yang, J.; Fan, D. Biotransformation of High Concentrations of Ginsenoside Substrate into Compound K by β-glycosidase from Sulfolobus solfataricus. Genes 2023, 14, 897. [Google Scholar] [CrossRef]
- Cui, C.-h.; Jeon, B.-M.; Fu, Y.; Im, W.-T.; Kim, S.-C. High-density Immobilization of a Ginsenoside-transforming β-glucosidase for Enhanced Food-grade Production of Minor ginsenosides. Appl. Microbiol. Biotechnol. 2019, 103, 7003–7015. [Google Scholar] [CrossRef]
- Sharma, A.; Lee, H.-J. Ginsenoside Compound K: Insights into Recent Studies on Pharmacokinetics and Health-promoting Activities. Biomolecules 2020, 10, 1028. [Google Scholar] [CrossRef]
- Yang, X.-D.; Yang, Y.-Y.; Ouyang, D.-S.; Yang, G.-P. A Review of Biotransformation and Pharmacology of Ginsenoside Compound K. Fitoterapia 2015, 100, 208–220. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Y.; Yu, Z.; Lv, G.; Huang, X.; Lin, H.; Ma, C.; Lin, Z.; Qu, P. Functional Mechanism of Ginsenoside Compound K on Tumor Growth and Metastasis. Integr. Cancer Ther. 2022, 21, 1–13. [Google Scholar] [CrossRef]
- Chen, L.; Zhou, L.; Huang, J.; Wang, Y.; Yang, G.; Tan, Z.; Wang, Y.; Zhou, G.; Liao, J.; Ouyang, D. Single- and Multiple-Dose Trials to Determine the Pharmacokinetics, Safety, Tolerability, and Sex Effect of Oral Ginsenoside Compound K in Healthy Chinese Volunteers. Front. Pharmacol. 2018, 8, 965. [Google Scholar] [CrossRef]
- Oh, J.; Kim, J.-S. Compound K Derived from Ginseng: Neuroprotection and Cognitive Improvement. Food Funct. 2016, 7, 4506–4515. [Google Scholar] [CrossRef]
- Lu, C.; Yin, Y. Pulsed Electric Field Treatment Combined With Commercial Enzymes Converts Major Ginsenoside Rb1 to Minor Ginsenoside Rd. Innov. Food Sci. Emerg. Technol. 2014, 22, 95–101. [Google Scholar] [CrossRef]
- Zhong, F.-L.; Ma, R.; Jiang, M.; Dong, W.-W.; Jiang, J.; Wu, S.; Li, D.; Quan, L.-H. Cloning and Characterization of Ginsenoside-hydrolyzing β-glucosidase from Lactobacillus brevis That Transforms Ginsenosides Rb1 and F2 into Ginsenoside Rd and Compound K. J. Microbiol. Biotechnol. 2016, 26, 1661–1667. [Google Scholar] [CrossRef]
- Kim, M.-J.; Upadhyaya, J.; Yoon, M.-S.; Ryu, N.S.; Song, Y.E.; Park, H.-W.; Kim, Y.-H.; Kim, M.-K. Highly Regioselective Biotransformation of Ginsenoside Rb2 into Compound Y and Compound K by β-glycosidase Purified from Armillaria mellea mycelia. J. Ginseng Res. 2018, 42, 504–511. [Google Scholar] [CrossRef]
- Kim, S.A.; Shin, K.C.; Oh, D.K. Complete Biotransformation of Protopanaxadiol-type Ginsenosides into 20-O-β-glucopyranosyl-20(S)-protopanaxadiol by Permeabilized Recombinant Escherichia coli Cells Coexpressing β-glucosidase and Chaperone Genes. J. Agric. Food Chem. 2019, 67, 8393–8401. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.C.; Kim, T.H.; Choi, J.H.; Oh, D.K. Complete Biotransformation of Protopanaxadiol-type Ginsenosides to 20- O-β-glucopyranosyl-20(S)-protopanaxadiol Using a Novel and Thermostable β-Glucosidase. J. Agric. Food Chem. 2018, 66, 2822–2829. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.N.A.; Son, J.S.; Awais, M.; Ko, J.H.; Yang, D.C.; Jung, S.K. β-glucosidase and Its Application in Bioconversion of Ginsenosides in Panax ginseng. Bioengineering 2023, 10, 484. [Google Scholar] [CrossRef]
- Kim, S.Y.; Lee, H.N.; Hong, S.J.; Kang, H.J.; Cho, J.Y.; Kim, D.; Ameer, K.; Kim, Y.M. Enhanced Biotransformation of the Minor Ginsenosides in Red Ginseng Extract by Penicillium decumbens β-glucosidase. Enzym. Microb. Technol. 2022, 153, 109941. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Zhu, C.; Shi, J.; Fan, D.; Deng, J.; Fu, R.; Huang, R.; Fan, C. High Efficiency Production of Ginsenoside Compound K by Catalyzing Ginsenoside Rb1 Using Snailase. Chin. J. Chem. Eng. 2018, 26, 1591–1597. [Google Scholar] [CrossRef]
- Yan, Q.; Zhou, W.; Shi, X.; Zhou, P.; Ju, D.; Feng, M. Biotransformation Pathways of Ginsenoside Rb1 to Compound K by β-glucosidases in Fungus Paecilomyces Bainier sp. 229. Process Biochem. 2010, 45, 1550–1556. [Google Scholar] [CrossRef]
- Kim, D.; Ku, S. Bacillus Cellulase Molecular Cloning, Expression, and Surface Display on the Outer Membrane of Escherichia coli. Molecules 2018, 23, 503. [Google Scholar] [CrossRef]
- Zhou, R.; Dong, S.; Feng, Y.; Cui, Q.; Xuan, J. Development of Highly Efficient Whole-cell Catalysts of Cis-epoxysuccinic Acid Hydrolase by Surface Display. Bioresour. Bioprocess. 2022, 9, 92. [Google Scholar] [CrossRef]
- Han, L.; Zhao, Y.; Cui, S.; Liang, B. Redesigning of Microbial Cell Surface and Its Application to Whole-cell Biocatalysis and Biosensors. Appl. Biochem. Biotechnol. 2017, 185, 396–418. [Google Scholar] [CrossRef]
- Suzuki, H.; Thongbhubate, K.; Muraoka, M.; Sasabu, A. Agmatine Production by Escherichia coli Cells Expressing SpeA on the Extracellular Surface. Enzym. Microb. Technol. 2023, 162, 110139. [Google Scholar] [CrossRef]
- Kondo, A.; Tanaka, T.; Hasunuma, T.; Ogino, C. Applications of Yeast Cell-surface Display in Bio-refinery. Recent Pat. Biotechnol. 2010, 4, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, K.; Ueda, M. Cell Surface Engineering of Yeast for Applications in White Biotechnology. Biotechnol. Lett. 2011, 33, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Narita, J.; Okano, K.; Tateno, T.; Tanino, T.; Sewaki, T.; Sung, M.-H.; Fukuda, H.; Kondo, A. Display of Active Enzymes on the Cell Surface of Escherichia coli Using PgsA Anchor Protein and Their Application to Bioconversion. Appl. Microbiol. Biotechnol. 2005, 70, 564–572. [Google Scholar] [CrossRef]
- Lee, S.Y.; Choi, J.H.; Xu, Z.H. Microbial Cell-surface Display. Trends Biotechnol. 2003, 21, 45–52. [Google Scholar] [CrossRef]
- Nakatani, H.; Hori, K. Cell Surface Protein Engineering for High-performance Whole-cell Catalysts. Front. Chem. Sci. Eng. 2017, 11, 46–57. [Google Scholar] [CrossRef]
- Georgiou, G.; Stephens, D.L.; Stathopoulos, C.; Poetschke, H.L.; Mendenhall, J.; Earhart, C.F. Display of Beta-lactamase on the Escherichia coli Surface: Outer Membrane Phenotypes Conferred by Lpp’-OmpA’-beta-lactamase Fusions. Protein Eng. 1996, 9, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Han, M.J. Novel Bacterial Surface Display System Based on the Escherichia coli Protein MipA. J. Microbiol. Biotechnol. 2020, 30, 1097–1103. [Google Scholar] [CrossRef]
- Han, M.J.; Lee, S.H. An Efficient Bacterial Surface Display System Based on a Novel Outer Membrane Anchoring Element from the Escherichia coli Protein YiaT. FEMS Microbiol. Lett. 2015, 362, 1–7. [Google Scholar] [CrossRef]
- Quan, L.H.; Min, J.W.; Jin, Y.; Wang, C.; Kim, Y.J.; Yang, D.C. Enzymatic Biotransformation of Ginsenoside Rb1 to Compound K by Recombinant β-glucosidase from Microbacterium esteraromaticum. J. Agric. Food Chem. 2012, 60, 3776–3781. [Google Scholar] [CrossRef]
- Maurer, J.; Jose, J.; Meyer, T. Autodisplay: One-component System for Efficient Surface Display and Release of Soluble Recombinant Proteins from Escherichia coli. J. Bacteriol. 1997, 179, 794–804. [Google Scholar] [CrossRef]
- Xia, H.; Li, N.; Zhong, X.; Jiang, Y. Metal-organic Frameworks: A Potential Platform for Enzyme Immobilization and Related Applications. Front. Bioeng. Biotechnol. 2020, 8, 695. [Google Scholar] [CrossRef] [PubMed]
- Donovan, R.S.; Robinson, C.W.; Glick, B.R. Review: Optimizing Inducer and Culture Conditions for Expression of Foreign Proteins Under the Control of the lac Promoter. J. Ind. Microbiol. 1996, 16, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Zarei Jaliani, H.; Farajnia, S.; Safdari, Y.; Mohammadi, S.A.; Barzegar, A.; Talebi, S. Optimized Condition for Enhanced Soluble-expression of Recombinant Mutant Anabaena Variabilis Phenylalanine Ammonia Lyase. Adv. Pharm. Bull. 2014, 4, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Bhatwa, A.; Wang, W.; Hassan, Y.I.; Abraham, N.; Li, X.Z.; Zhou, T. Challenges Associated With the Formation of Recombinant Protein Inclusion Bodies in Escherichia coli and Strategies to Address Them for Industrial Applications. Front. Bioeng. Biotechnol. 2021, 9, 630551. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Ou, L.; Wei, D.; Wei, X. Optimum Induction of Recombinant Thymidine Phosphorylase and Its Application. Nucleosides Nucleotides Nucleic Acids 2011, 30, 360–368. [Google Scholar] [CrossRef]
- Chen, X.; Li, C.; Liu, H. Enhanced Recombinant Protein Production under Special Environmental Stress. Front. Microbiol. 2021, 12, 630814. [Google Scholar] [CrossRef]
- Juneja, V.K.; Mukhopadhyay, S.; Ukuku, D.; Hwang, C.A.; Wu, V.C.; Thippareddi, H. Interactive Effects of Temperature, pH, and Water Activity on the Growth Kinetics of Shiga Toxin-producing Escherichia coli O104:H4. J. Food Prot. 2014, 77, 706–712. [Google Scholar] [CrossRef]
- Cabilly, S. Growth at Sub-optimal Temperatures Allows the Production of Functional, Antigen-binding Fab Fragments in Escherichia coli. Gene 1989, 85, 553–557. [Google Scholar] [CrossRef]
- Huang, R.; Zhang, F.; Yan, X.; Qin, Y.; Jiang, J.; Liu, Y.; Song, Y. Characterization of the β-glucosidase Activity in Indigenous Yeast Isolated from Wine Regions in China. J. Food Sci. 2021, 86, 2327–2345. [Google Scholar] [CrossRef]








| Type | Specific Enzyme Activity (U/mg) | Ginsenoside CK Production (mg/mL) | Conversion (%) |
|---|---|---|---|
| Whole cell (pET28a-yiaT-bgp3) | 57.06 ± 0.95 | 1.07 ± 0.05 | 50.48 ± 2.34 |
| Lysate (pET28a-bgp3) | 56.10 ± 1.45 | 1.06 ± 0.07 | 50.24 ± 3.35 |
| Whole cell (pET28a-bgp3) | 10.91 ± 1.26 | 0.31 ± 0.04 | 14.69 ± 2.01 |
| Strains or Plasmids | Descriptions | Source |
|---|---|---|
| E. coli DH5α | Clone strain | Vazyme |
| E. coli BL21(DE3) | Expression strain | Vazyme |
| pET28a | pBR232 origin, lac I coding sequence, PT7, Kmr | Lab stock |
| pET28a-bgp3 | pET28a derivative, PT7, Kmr, intracellular Bgp3 expression | This study |
| pET28a-yiaT-bgp3 | pET28a derivative, PT7, Kmr, fusion expression of YiaT-Bgp3 | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, L.; Li, T.; Guo, G.; Liu, Z.; Hao, N. The Synthesis of Ginsenoside Compound K Using a Surface-Displayed β-Glycosidase Whole-Cell Catalyst. Catalysts 2023, 13, 1375. https://doi.org/10.3390/catal13101375
Guo L, Li T, Guo G, Liu Z, Hao N. The Synthesis of Ginsenoside Compound K Using a Surface-Displayed β-Glycosidase Whole-Cell Catalyst. Catalysts. 2023; 13(10):1375. https://doi.org/10.3390/catal13101375
Chicago/Turabian StyleGuo, Lianxia, Tao Li, Gege Guo, Zhaoxing Liu, and Ning Hao. 2023. "The Synthesis of Ginsenoside Compound K Using a Surface-Displayed β-Glycosidase Whole-Cell Catalyst" Catalysts 13, no. 10: 1375. https://doi.org/10.3390/catal13101375