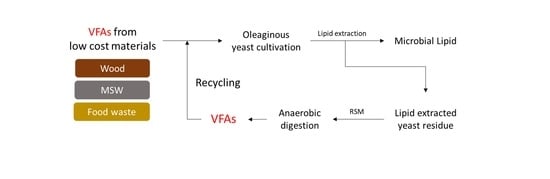
Volatile Fatty Acids from Lipid-Extracted Yeast Provide Additional Feedstock for Microbial Lipid Production
Abstract
:1. Introduction
2. Results and Discussion
2.1. Preparation of a LEY Residue
2.2. Application of Response Surface Methodology for the Optimization of VFA Production via Anaerobic Digestion
2.3. Recycling of the LEY Residue as a Feedstock for VFA Production
3. Materials and Methods
3.1. Yeast Cultivation
3.2. Lipid Extraction of Yeast and Preparation LEY Residue Sample
3.3. Anaerobic Digestion Operating Conditions
3.4. VFA Concentration Analysis
3.5. Central Composite in Cube in Design and Selection of Variables
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Papanikolaou, S.; Aggelis, G. Lipids of oleaginous yeasts. Part I: Biochemistry of single cell oil production. Eur. J. Lipid Sci. Technol. 2011, 113, 1031–1051. [Google Scholar] [CrossRef]
- Wynn, J.; Ratledge, C. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv. Appl. Microbiol. 2002, 51, 1–51. [Google Scholar]
- Fakas, S.; Papanikolaou, S.; Galiotou-Panayotou, M.; Komaitis, M.; Aggelis, G. Organic nitrogen of tomato waste hydrolysate enhances glucose uptake and lipid accumulation in Cunninghamella echinulata. J. Appl. Microbiol. 2008, 105, 1062–1070. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Aggelis, G. Yarrowia lipolytica: A model microorganism used for the production of tailor-made lipids. Eur. J. Lipid Sci. Technol. 2010, 112, 639–654. [Google Scholar] [CrossRef]
- FAO. The State of Food and Agriculture, Biofuels: Prospects, Risks and Opportunities; Food and Agriculture Organization of the United Nations: Rome, Italy, 2008. [Google Scholar]
- Chisti, Y. Biodiesel from microalgae. Biotechnol. Adv. 2007, 25, 294–306. [Google Scholar] [CrossRef]
- Fei, Q.; Chang, H.N.; Shang, L.; Choi, J.-D.-R. Exploring low-cost carbon sources for microbial lipids production by fed-batch cultivation of Cryptococcus albidus. Biotechnol. Bioprocess Eng. 2011, 16, 482–487. [Google Scholar] [CrossRef]
- Sathiyamoorthi, E.; Kumar, P.; Kim, B.S. Lipid production by Cryptococcus albidus using biowastes hydrolysed by indigenous microbes. Bioprocess Biosyst. Eng. 2019, 42, 687–696. [Google Scholar] [CrossRef]
- Park, G.W.; Fei, Q.; Jung, K.; Chang, H.N.; Kim, Y.-C.; Kim, N.j.; Choi, J.d.R.; Kim, S.; Cho, J. Volatile fatty acids derived from waste organics provide an economical carbon source for microbial lipids/biodiesel production. Biotechnol. J. 2014, 9, 1536–1546. [Google Scholar] [CrossRef]
- Huang, X.; Chen, R.; Yuan, M.; Liu, J. Efficient bioconversion of high-content volatile fatty acids into microbial lipids by Cryptococcus curvatus ATCC 20509. Bioresour. Technol. 2017, 239, 394–401. [Google Scholar]
- Liu, J.; Yuan, M.; Liu, J.-N.; Huang, X.-F. Bioconversion of mixed volatile fatty acids into microbial lipids by Cryptococcus curvatus ATCC 20509. Bioresour. Technol. 2017, 241, 645–651. [Google Scholar] [CrossRef]
- Park, G.W.; Chang, H.N.; Jung, K.; Seo, C.; Kim, Y.-C.; Choi, J.H.; Woo, H.C.; Hwang, I.-J. Production of microbial lipid by Cryptococcus curvatus on rice straw hydrolysates. Process. Biochem. 2017, 56, 147–153. [Google Scholar] [CrossRef]
- Fontanille, P.; Kumar, V.; Christophe, G.; Nouaille, R.; Larroche, C. Bioconversion of volatile fatty acids into lipids by the oleaginous yeast Yarrowia lipolytica. Bioresour. Technol. 2012, 114, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Llamas, M.; Tomás-Pejó, E.; González-Fernández, C. Volatile Fatty Acids from Organic Wastes as Novel Low-cost carbon source for Yarrowia lipolytica. New Biotechnol. 2020, 56, 123–129. [Google Scholar] [CrossRef]
- Huang, X.-F.; Liu, J.-N.; Lu, L.-J.; Peng, K.-M.; Yang, G.-X.; Liu, J. Culture strategies for lipid production using acetic acid as sole carbon source by Rhodosporidium toruloides. Bioresour. Technol. 2016, 206, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Fei, Q.; O’Brien, M.; Nelson, R.; Chen, X.; Lowell, A.; Dowe, N. Enhanced lipid production by Rhodosporidium toruloides using different fed-batch feeding strategies with lignocellulosic hydrolysate as the sole carbon source. Biotechnol. Biofuels 2016, 9, 130. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.N.; Kim, N.-J.; Kang, J.; Jeong, C.M. Biomass-derived volatile fatty acid platform for fuels and chemicals. Biotechnol. Bioprocess Eng. 2010, 15, 1–10. [Google Scholar] [CrossRef]
- Park, G.W.; Seo, C.; Jung, K.; Chang, H.N.; Kim, W.; Kim, Y.-C. A comprehensive study on volatile fatty acids production from rice straw coupled with microbial community analysis. Bioprocess Biosyst. Eng. 2015, 38, 1157–1166. [Google Scholar] [CrossRef]
- Zheng, Y.; Chi, Z.; Ahring, B.K.; Chen, S. Oleaginous yeast Cryptococcus curvatus for biofuel production: Ammonia’s effect. Biomass Bioenergy 2012, 37, 114–121. [Google Scholar] [CrossRef]
- Sri Bala Kameswari, K.; Chitra, K.; Porselvam, S.; Thanasekaran, K. Optimization of inoculum to substrate ratio for bio-energy generation in co-digestion of tannery solid wastes. Clean Technol. Environ. Policy 2011, 14, 241–250. [Google Scholar] [CrossRef]
- Pham, T.N.; Nam, W.J.; Jeon, Y.J.; Yoon, H.H. Volatile fatty acids production from marine macroalgae by anaerobic fermentation. Bioresour. Technol. 2012, 124, 500–503. [Google Scholar] [CrossRef]
- Pan, J.; Zhang, R.; Elmashad, H.; Sun, H.; Ying, Y. Effect of food to microorganism ratio on biohydrogen production from food waste via anaerobic fermentation. Int. J. Hydrogen Energy 2008, 33, 6968–6975. [Google Scholar] [CrossRef]
- Gebremariam, S.; Marchetti, J.M. Economics of biodiesel production. Energy Convers. Manag. 2018, 168, 74–84. [Google Scholar] [CrossRef]
- Meesters, P.; Huijberts, G.; Eggink, G. High-cell-density cultivation of the lipid accumulating yeast Cryptococcus curvatus using glycerol as a carbon source. Appl. Microbiol. Biotechnol. 1996, 45, 575–579. [Google Scholar] [CrossRef]
- Folch, J.; Lees, M.; Sloane-Stanley, G. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [Google Scholar] [CrossRef]
- Yang, K.; Oh, C.; Hwang, S. Optimizing volatile fatty acid production in partial acidogenesis of swine wastewater. Water Sci. Technol. 2004, 50, 169–176. [Google Scholar] [CrossRef] [PubMed]



| Run | pH | Inoculation (%, v/v) | VFA Concentration (g/L) |
|---|---|---|---|
| 1 | 6.5 | 5 | 3.10 |
| 2 | 6.5 | 25 | 3.75 |
| 3 | 6.5 | 45 | 3.41 |
| 4 | 9.0 | 5 | 3.82 |
| 5 * | 9.0 | 25 | 4.20 |
| 6 * | 9.0 | 25 | 4.51 |
| 7 * | 9.0 | 25 | 4.27 |
| 8 | 9.0 | 45 | 4.68 |
| 9 | 11.5 | 5 | 1.38 |
| 10 | 11.5 | 25 | 1.75 |
| 11 | 11.5 | 45 | 1.95 |
| Source | Degrees of Freedom | VFA Concentration | |
|---|---|---|---|
| Mean Square | p-Value | ||
| Model | 5 | 2.67 | 0.0001 |
| X1 | 1 | 4.45 | 0.0001 |
| X2 | 1 | 0.51 | 0.0151 |
| X1X2 | 1 | 0.018 | 0.5243 |
| X12 | 1 | 7.26 | <0.0001 |
| X22 | 1 | 0.091 | 0.1863 |
| Residual | 5 | 0.039 | |
| Lack of fit | 3 | 0.047 | 0.3782 |
| Pure error | 2 | 0.026 | |
| Correlation total | 10 | ||
| R2 = 97.15% for VFA concentration | |||
| Factor | Coded Levels | ||
|---|---|---|---|
| −1 | 0 | 1 | |
| pH, X1 | 6.5 | 9.0 | 11.5 |
| Inoculation (%), X2 | 5 | 25 | 45 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, G.W.; Son, S.; Moon, M.; Sin, S.; Min, K.; Lee, J.-S.; Chang, H.N. Volatile Fatty Acids from Lipid-Extracted Yeast Provide Additional Feedstock for Microbial Lipid Production. Catalysts 2021, 11, 1009. https://doi.org/10.3390/catal11081009
Park GW, Son S, Moon M, Sin S, Min K, Lee J-S, Chang HN. Volatile Fatty Acids from Lipid-Extracted Yeast Provide Additional Feedstock for Microbial Lipid Production. Catalysts. 2021; 11(8):1009. https://doi.org/10.3390/catal11081009
Chicago/Turabian StylePark, Gwon Woo, Seongsoo Son, Myounghoon Moon, Subin Sin, Kyoungseon Min, Jin-Suk Lee, and Ho Nam Chang. 2021. "Volatile Fatty Acids from Lipid-Extracted Yeast Provide Additional Feedstock for Microbial Lipid Production" Catalysts 11, no. 8: 1009. https://doi.org/10.3390/catal11081009