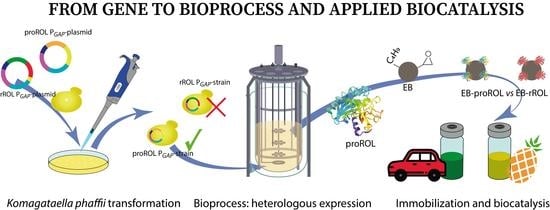
Constitutive Expression in Komagataella phaffii of Mature Rhizopus oryzae Lipase Jointly with Its Truncated Prosequence Improves Production and the Biocatalyst Operational Stability
Abstract
:1. Introduction
2. Results and Discussion
2.1. Batch and Fed-Batch Bioprocess Strategies. Lipase Production under Inducible PAOX1 and Constitutive PGAP
2.2. N-Terminal Amino Acids in proROL, rROL and proROLm Free Enzymes
2.3. Stability of EB-proROL and EB-rROL in the Presence of Organic Solvents
2.4. Transesterification and Esterification Reactions. Initial Reaction Rate and Operational Stability
2.4.1. Biodiesel Production
2.4.2. Ethyl Butyrate Production
3. Materials and Methods
3.1. Chemicals
3.2. Strains and Plasmids
3.3. Batch Cultures
3.4. Fed-Batch Cultures
3.5. Lipase Production and Activity Assessment
3.6. N-Terminal Analysis
3.7. Lipase Immobilization
3.8. Transesterification and Esterification Reactions
3.9. Gas Chromatography
3.10. Immobilized Lipase Stability against Organic Solvents
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ROL | Rhizopus oryzae lipase. |
| rROL | Recombinant Rhizopus oryzae lipase formed by the mature sequence. |
| proROL | Lipase formed by the C-terminal 28 amino acids of the native prosequence of ROL fused to the N-terminal in the mature sequence. |
| proROLm | proROL modified by the activity of proteases. |
| w-proROL-gene: Gene encoding the whole prosequece fused to the mature sequence of Rhizopus oryzae lipase. | |
| rROL-gene | Gene encoding the mature sequence of Rhizopus oryzae lipase. |
| proROL-gene | Gene encoding the C-terminal 28 amino acids of the native prosequence of ROL fused to the N-terminal of the mature sequence. |
| EB | Purolite® polymethacrylate matrix support with butyl and epoxide surface groups |
| EB-rROL | rROL covalently immobilized onto the EB support. |
| EB-proROL | proROL covalently immobilized onto the EB support. |
| rROL PAOX1-plasmid | Plasmid containing the mature sequence of Rhizopus oryzae lipase under PAOX1. |
| proROL PAOX1-plasmid | Plasmid containing the sequence of proROL under PAOX1. |
| rROL PGAP-plasmid | Plasmid containing the mature sequence of Rhizopus oryzae lipase under PGAP. |
| proROL PGAP-plasmid | Plasmid containing the proROL sequence under PGAP. |
| rROL PAOX1-strain | Genetically modified Komagataella phaffii strain used to produce rROL under PAOX1. |
| proROL PAOX1-strain | Genetically modified Komagataella phaffii strain used to produce proROL under PAOX1. |
| rROL PGAP-strain | Genetically modified Komagataella phaffii strain used to produce rROL under PGAP. |
| proROL PGAP-strain | Genetically modified Komagataella phaffii strain used to produce proROL under PGAP. |
| MNLFB | Methanol non-limited fed-batch. |
| MLFB | Methanol limited fed-batch. |
| Y(P/X) | Product biomass yield (AU gX−1). |
| µ | Specific growth rate (h−1). |
| qp | Specific production rate (AU gX−1 h−1): parameter which shows an average of all the analyzed points during the bioprocess. |
| Specific productivity (AU gX−1 h−1) | Parameter of the entire bioprocess, analyzed at the end of it. |
| Volumetric productivity (AU L−1 h−1) | Parameter of the entire bioprocess, analyzed at the end of it. |
| PAOX1 | Inducible alcohol oxidase 1 promoter. |
| PGAP | Constitutive glyceraldehyde-3-phosphate dehydrogenase |
References
- Bornscheuer, U.T. The fourth wave of biocatalysis is approaching. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2018, 376, 20170063. [Google Scholar] [CrossRef] [PubMed]
- Bilal, M.; Iqbal, H.M.N. Armoring bio-catalysis via structural and functional coordination between nanostructured materials and lipases for tailored applications. Int. J. Biol. Macromol. 2021, 166, 818–838. [Google Scholar] [CrossRef] [PubMed]
- Todo Bom, M.A.; Botton, V.; Altheia, F.M.; Thomas, J.C.; Piovan, L.; Córdova, J.; Mitchell, D.A.; Krieger, N. Fermented solids that contain lipases produced by Rhizopus microsporus have an S-enantiopreference in the resolution of secondary alcohols. Biochem. Eng. J. 2021, 165, 107817. [Google Scholar] [CrossRef]
- Ismail, A.R.; Kashtoh, H.; Baek, K.H. Temperature-resistant and solvent-tolerant lipases as industrial biocatalysts: Biotechnological approaches and applications. Int. J. Biol. Macromol. 2021, 187, 127–142. [Google Scholar] [CrossRef]
- López-Fernández, J.; Benaiges, M.D.; Valero, F. Rhizopus oryzae lipase, a promising industrial enzyme: Biochemical characteristics, production and biocatalytic applications. Catalysts 2020, 10, 1277. [Google Scholar] [CrossRef]
- Salah, R.B.; Mosbah, H.; Fendri, A.; Gargouri, A.; Gargouri, Y.; Mejdoub, H. Biochemical and molecular characterization of a lipase produced by Rhizopus oryzae. FEMS Microbiol. Lett. 2006, 260, 241–248. [Google Scholar] [CrossRef] [Green Version]
- López-Fernández, J.; Barrero, J.J.; Benaiges, M.D.; Valero, F. Truncated prosequence of Rhizopus oryzae lipase: Key factor for production improvement and biocatalyst stability. Catalysts 2019, 9, 961. [Google Scholar] [CrossRef] [Green Version]
- Resina, D.; Maurer, M.; Cos, O.; Arnau, C.; Carnicer, M.; Marx, H.; Gasser, B.; Valero, F.; Mattanovich, D.; Ferrer, P. Engineering of bottlenecks in Rhizopus oryzae lipase production in Pichia pastoris using the nitrogen source-regulated FLD1 promoter. New Biotechnol. 2009, 25, 396–403. [Google Scholar] [CrossRef]
- Satomura, A.; Kuroda, K.; Ueda, M. Generation of a functionally distinct Rhizopus oryzae lipase through protein folding memory. PLoS ONE 2015, 10, e0124545. [Google Scholar] [CrossRef] [Green Version]
- Beer, H.D.; Wohlfahrt, G.; Schmid, R.D.; McCarthy, J.E. The folding and activity of the extracellular lipase of Rhizopus oryzae are modulated by a prosequence. Biochem. J. 1996, 319, 351–359. [Google Scholar] [CrossRef] [Green Version]
- Shinde, U.; Inouye, M. Intramolecular chaperones: Polypeptide extensions that modulate protein folding. Semin. Cell Dev. Biol. 2000, 11, 35–44. [Google Scholar] [CrossRef]
- Chapman, J.; Ismail, A.E.; Dinu, C.Z. Industrial applications of enzymes: Recent advances, techniques, and outlooks. Catalysts 2018, 8, 238. [Google Scholar] [CrossRef] [Green Version]
- Bilal, M.; Cui, J.; Iqbal, H.M.N. Tailoring enzyme microenvironment: State-of-the-art strategy to fulfill the quest for efficient bio-catalysis. Int. J. Biol. Macromol. 2019, 130, 186–196. [Google Scholar] [CrossRef]
- Bilal, M.; Iqbal, H.M.N.; Guo, S.; Hu, H.; Wang, W.; Zhang, X. State-of-the-art protein engineering approaches using biological macromolecules: A review from immobilization to implementation view point. Int. J. Biol. Macromol. 2018, 108, 893–901. [Google Scholar] [CrossRef]
- Ismail, A.R.; Baek, K.H. Lipase immobilization with support materials, preparation techniques, and applications: Present and future aspects. Int. J. Biol. Macromol. 2020, 163, 1624–1639. [Google Scholar] [CrossRef]
- Bonet-Ragel, K.; López-Pou, L.; Tutusaus, G.; Benaiges, M.D.; Valero, F. Rice husk ash as a potential carrier for the immobilization of lipases applied in the enzymatic production of biodiesel. Biocatal. Biotransform. 2018, 36, 151–158. [Google Scholar] [CrossRef]
- Pashangeh, K.; Akhond, M.; Karbalaei-Heidari, H.R.; Absalan, G. Biochemical characterization and stability assessment of Rhizopus oryzae lipase covalently immobilized on amino-functionalized magnetic nanoparticles. Int. J. Biol. Macromol. 2017, 105, 300–307. [Google Scholar] [CrossRef]
- Ghattas, N.; Abidi, F.; Galai, S.; Marzouki, M.N.; Salah, A. Ben Monoolein production by triglycerides hydrolysis using immobilized Rhizopus oryzae lipase. Int. J. Biol. Macromol. 2014, 68, 1–6. [Google Scholar] [CrossRef]
- Kartal, F.; Kilinc, A. Crosslinked aggregates of Rhizopus oryzae lipase as industrial biocatalysts: Preparation, optimization, characterization, and application for enantioselective resolution reactions. Biotechnol. Prog. 2012, 28, 937–945. [Google Scholar] [CrossRef]
- Yu, X.W.; Xu, Y.; Xiao, R. Lipases from the genus Rhizopus: Characteristics, expression, protein engineering and application. Prog. Lipid Res. 2016, 64, 57–68. [Google Scholar] [CrossRef]
- Beer, H.D.; McCarthy, J.E.G.; Bornscheuer, U.T.; Schmid, R.D. Cloning, expression, characterization and role of the leader sequence of a lipase from Rhizopus oryzae. Biochim. Biophys. Acta-Gene Struct. Expr. 1998, 1399, 173–180. [Google Scholar] [CrossRef]
- Di Lorenzo, M.; Hidalgo, A.; Haas, M.; Bornscheuer, U.T. Heterologous production of functional forms of Rhizopus oryzae lipase in Escherichia coli. Appl. Environ. Microbiol. 2005, 71, 8974–8977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, W.; Li, Z.; Tan, T. Secretion of pro- and mature Rhizopus arrhizus lipases by Pichia pastoris and properties of the proteins. Mol. Biotechnol. 2006, 32, 73–81. [Google Scholar] [CrossRef]
- Minning, S.; Schmidt-Dannert, C.; Schmid, R.D. Functional expression of Rhizopus oryzae lipase in Pichia pastoris: High-level production and some properties. J. Biotechnol. 1998, 66, 147–156. [Google Scholar] [CrossRef]
- Chahed, H.; Boumaiza, M.; Ezzine, A.; Marzouki, M.N. Heterologous expression and biochemical characterization of a novel thermostable Sclerotinia sclerotiorum GH45 endoglucanase in Pichia pastoris. Int. J. Biol. Macromol. 2018, 106, 629–635. [Google Scholar] [CrossRef]
- Sturmberger, L.; Chappell, T.; Geier, M.; Krainer, F.; Day, K.J.; Vide, U.; Trstenjak, S.; Schiefer, A.; Richardson, T.; Soriaga, L.; et al. Refined Pichia pastoris reference genome sequence. J. Biotechnol. 2016, 235, 121–131. [Google Scholar] [CrossRef] [Green Version]
- Garrigós-Martínez, J.; Nieto-Taype, M.A.; Gasset-Franch, A.; Montesinos-Seguí, J.L.; Garcia-Ortega, X.; Valero, F. Specific growth rate governs AOX1 gene expression, affecting the production kinetics of Pichia pastoris (Komagataella phaffii) PAOX1-driven recombinant producer strains with different target gene dosage. Microb. Cell Fact. 2019, 18, 187. [Google Scholar] [CrossRef]
- Garrigós-Martínez, J.; Vuoristo, K.; Nieto-Taype, M.A.; Tähtiharju, J.; Uusitalo, J.; Tukiainen, P.; Schmid, C.; Tolstorukov, I.; Madden, K.; Penttilä, M.; et al. Bioprocess performance analysis of novel methanol-independent promoters for recombinant protein production with Pichia pastoris. Microb. Cell Fact. 2021, 20, 74. [Google Scholar] [CrossRef]
- García-Ortega, X.; Cámara, E.; Ferrer, P.; Albiol, J.; Montesinos-Seguí, J.L.; Valero, F. Rational development of bioprocess engineering strategies for recombinant protein production in Pichia pastoris (Komagataella phaffii) using the methanol-free GAP promoter. Where do we stand? New Biotechnol. 2019, 53, 24–34. [Google Scholar] [CrossRef]
- Müller, J.M.; Bruhn, S.; Flaschel, E.; Friehs, K.; Risse, J.M. GAP promoter-based fed-batch production of highly bioactive core streptavidin by Pichia pastoris. Biotechnol. Prog. 2016, 32, 855–864. [Google Scholar] [CrossRef]
- Nieto-Taype, M.A.; Garrigós-Martínez, J.; Sánchez-Farrando, M.; Valero, F.; Garcia-Ortega, X.; Montesinos-Seguí, J.L. Rationale-based selection of optimal operating strategies and gene dosage impact on recombinant protein production in Komagataella phaffii (Pichia pastoris). Microb. Biotechnol. 2020, 13, 315–327. [Google Scholar] [CrossRef]
- Yu, X.W.; Yang, M.; Jiang, C.; Zhang, X.; Xu, Y. N-Glycosylation engineering to improve the constitutive expression of Rhizopus oryzae lipase in Komagataella phaffii. J. Agric. Food Chem. 2017, 65, 6009–6015. [Google Scholar] [CrossRef]
- Ponte, X.; Barrigón, J.M.; Maurer, M.; Mattanovich, D.; Valero, F.; Montesinos-Seguí, J.L. Towards optimal substrate feeding for heterologous protein production in Pichia pastoris (Komagataella spp.) fed-batch processes under PAOX1 control: A modeling aided approach. J. Chem. Technol. Biotechnol. 2018, 93, 3208–3218. [Google Scholar] [CrossRef]
- Kluge, J.; Terfehr, D.; Kück, U. Inducible promoters and functional genomic approaches for the genetic engineering of filamentous fungi. Appl. Microbiol. Biotechnol. 2018, 102, 6357–6372. [Google Scholar] [CrossRef] [Green Version]
- Hama, S.; Tamalampudi, S.; Shindo, N.; Numata, T.; Yamaji, H.; Fukuda, H.; Kondo, A. Role of N-terminal 28 amino acid region of Rhizopus oryzae lipase in directing proteins to secretory pathway of Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2008, 79, 1009–1018. [Google Scholar] [CrossRef]
- Barrigón, J.M.; Montesinos, J.L.; Valero, F. Searching the best operational strategies for Rhizopus oryzae lipase production in Pichia pastoris Mut+ phenotype: Methanol limited or methanol non-limited fed-batch cultures? Biochem. Eng. J. 2013, 75, 47–54. [Google Scholar] [CrossRef]
- Kohno, M.; Kugimiya, W.; Hashimoto, Y.; Morita, Y. Purification, characterization, and crystallization of two types of lipase from Rhizopus niveus. Biosci. Biotechnol. Biochem. 1994, 58, 1007–1012. [Google Scholar] [CrossRef]
- Guillén, M.; Benaiges, M.D.; Valero, F. Comparison of the biochemical properties of a recombinant lipase extract from Rhizopus oryzae expressed in Pichia pastoris with a native extract. Biochem. Eng. J. 2011, 54, 117–123. [Google Scholar] [CrossRef]
- Takahashi, S.; Ueda, M.; Tanaka, A. Independent production of two molecular forms of a recombinant Rhizopus oryzae lipase by KEX2-engineered strains of Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 1999, 52, 534–540. [Google Scholar] [CrossRef]
- Kumar, A.; Dhar, K.; Kanwar, S.S.; Arora, P.K. Lipase catalysis in organic solvents: Advantages and applications. Biol. Proced. Online 2016, 18, 2. [Google Scholar] [CrossRef] [Green Version]
- Hiol, A.; Jonzo, M.D.; Rugani, N.; Druet, D.; Sarda, L.; Comeau, L.C. Purification and characterization of an extracellular lipase from a thermophilic Rhizopus oryzae strain isolated from palm fruit. Enzym. Microb. Technol. 2000, 26, 421–430. [Google Scholar] [CrossRef]
- McDowell, C.; Bazan, G.C. Organic solar cells processed from green solvents. Curr. Opin. Green Sustain. Chem. 2017, 5, 49–54. [Google Scholar] [CrossRef]
- Wang, S.; Meng, X.; Zhou, H.; Liu, Y.; Secundo, F.; Liu, Y. Enzyme stability and activity in non-aqueous reaction systems: A mini review. Catalysts 2016, 6, 32. [Google Scholar] [CrossRef] [Green Version]
- Guillén, M.; Benaiges, M.D.; Valero, F. Biosynthesis of ethyl butyrate by immobilized recombinant Rhizopus oryzae lipase expressed in Pichia pastoris. Biochem. Eng. J. 2012, 65, 1–9. [Google Scholar] [CrossRef]
- Zhong, L.; Feng, Y.; Wang, G.; Wang, Z.; Bilal, M.; Lv, H.; Jia, S.; Cui, J. Production and use of immobilized lipases in/on nanomaterials: A review from the waste to biodiesel production. Int. J. Biol. Macromol. 2020, 152, 207–222. [Google Scholar] [CrossRef]
- López-Fernández, J.; Dolors Benaiges, M.; Valero, F. Second- and third-generation biodiesel production with immobilised recombinant Rhizopus oryzae lipase: Influence of the support, substrate acidity and bioprocess scale-up. Bioresour. Technol. 2021, 334, 125233. [Google Scholar] [CrossRef]
- Rodrigues, A.; Bordado, J.C.; Dos Santos, R.G. Upgrading the glycerol from biodiesel production as a source of energy carriers and chemicals—A technological review for three chemical pathways. Energies 2017, 10, 1817. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Ge, X.; Cui, S.; Li, Y. Value-added processing of crude glycerol into chemicals and polymers. Bioresour. Technol. 2016, 215, 144–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayari, A.; Frikha, F.; Miled, N.; Mtibaa, H.; Ali, Y.B.; Verger, R.; Gargouri, Y. N-terminal peptide of Rhizopus oryzae lipase is important for its catalytic properties. FEBS Lett. 2005, 579, 976–982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.; Jiang, T.; Wu, Y.; Cui, L.; Qin, S.; He, B. Elucidation of lid open and orientation of lipase activated in interfacial activation by amphiphilic environment. Int. J. Biol. Macromol. 2018, 119, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Shi, H.; Gu, H.; Naka, G.; Ding, H.; Li, X.; Zhang, Y.; Hu, B.; Wang, F. Immobilization of Rhizopus oryzae ly6 onto loofah sponge as a whole-cell biocatalyst for biodiesel production. BioResources 2016, 11, 850–860. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, J.; Canet, A.; Rivera, I.; Osório, N.M.; Sandoval, G.; Valero, F.; Ferreira-Dias, S. Biodiesel production from crude Jatropha oil catalyzed by non-commercial immobilized heterologous Rhizopus oryzae and Carica papaya lipases. Bioresour. Technol. 2016, 213, 88–95. [Google Scholar] [CrossRef] [Green Version]
- Asmat, S.; Anwer, A.H.; Husain, Q. Immobilization of lipase onto novel constructed polydopamine grafted multiwalled carbon nanotube impregnated with magnetic cobalt and its application in synthesis of fruit flavours. Int. J. Biol. Macromol. 2019, 140, 484–495. [Google Scholar] [CrossRef]
- Guillén, M.; Benaiges, M.D.; Valero, F. Improved ethyl butyrate synthesis catalyzed by an immobilized recombinant Rhizopus oryzae lipase: A comprehensive statistical study by production, reaction rate and yield analysis. J. Mol. Catal. B Enzym. 2016, 133, S371–S376. [Google Scholar] [CrossRef]
- Barrero, J.J.; Casler, J.C.; Valero, F.; Ferrer, P.; Glick, B.S. An improved secretion signal enhances the secretion of model proteins from Pichia pastoris. Microb. Cell Fact. 2018, 17, 161–174. [Google Scholar] [CrossRef]
- Cámara, E.; Albiol, J.; Ferrer, P. Droplet digital PCR-aided screening and characterization of Pichia pastoris multiple gene copy strains. Biotechnol. Bioeng. 2016, 113, 1542–1551. [Google Scholar] [CrossRef]
- Maurer, M.; Kühleitner, M.; Gasser, B.; Mattanovich, D. Versatile modeling and optimization of fed batch processes for the production of secreted heterologous proteins with Pichia pastoris. Microb. Cell Fact. 2006, 5, 37–47. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Ortega, X.; Ferrer, P.; Montesinos, J.L.; Valero, F. Fed-batch operational strategies for recombinant Fab production with Pichia pastoris using the constitutive GAP promoter. Biochem. Eng. J. 2013, 79, 172–181. [Google Scholar] [CrossRef]
- Ponte, X.; Montesinos-Seguí, J.L.; Valero, F. Bioprocess efficiency in Rhizopus oryzae lipase production by Pichia pastoris under the control of PAOX1 is oxygen tension dependent. Process Biochem. 2016, 51, 1954–1963. [Google Scholar] [CrossRef]
- Mendes, A.A.; Oliveira, P.C.; Vélez, A.M.; Giordano, R.C.; de LC Giordano, R.; de Castro, H.F. Evaluation of immobilized lipases on poly-hydroxybutyrate beads to catalyze biodiesel synthesis. Int. J. Biol. Macromol. 2012, 50, 503–511. [Google Scholar] [CrossRef]
- Resina, D.; Serrano, A.; Valero, F.; Ferrer, P. Expression of a Rhizopus oryzae lipase in Pichia pastoris under control of the nitrogen source-regulated formaldehyde dehydrogenase promoter. J. Biotechnol. 2004, 109, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Romero, E.; Ferreira, P.; Martínez, Á.T.; Martínez, M.J. New oxidase from Bjerkandera arthroconidial anamorph that oxidizes both phenolic and nonphenolic benzyl alcohols. Biochim. Biophys. Acta-Proteins Proteom. 2009, 1794, 689–697. [Google Scholar] [CrossRef] [PubMed]
- Bonet-Ragel, K.; Canet, A.; Benaiges, M.D.; Valero, F. Synthesis of biodiesel from high FFA alperujo oil catalysed by immobilised lipase. Fuel 2015, 161, 12–17. [Google Scholar] [CrossRef]
- Bonet-Ragel, K.; Canet, A.; Benaiges, M.D.; Valero, F. Effect of acyl-acceptor stepwise addition strategy using alperujo oil as a substrate in enzymatic biodiesel synthesis. J. Chem. Technol. Biotechnol. 2018, 93, 541–547. [Google Scholar] [CrossRef] [Green Version]
- Aymard, C.; Belarbi, A. Kinetics of thermal deactivation of enzymes: A simple three parameters phenomenological model can describe the decay of enzyme activity, irrespectively of the mechanism. Enzym. Microb. Technol. 2000, 27, 612–618. [Google Scholar] [CrossRef]
- Canet, A.; Dolors Benaiges, M.; Valero, F. Biodiesel synthesis in a solvent-free system by recombinant Rhizopus oryzae lipase. Study of the catalytic reaction progress. JAOCS J. Am. Oil Chem. Soc. 2014, 91, 1499–1506. [Google Scholar] [CrossRef]



| Parameter | proROL PAOX1 1 | proROL PGAP | rROL PAOX1 1 |
|---|---|---|---|
| Final activity (AU mL−1) | 12.38 | 74.71 | 10.51 |
| Y(P/X) (total AU total gX−1) | 5017 | 4273 | 3753 |
| µ (h−1) | 0.073 | 0.22 | 0.045 |
| qp (AU gX−1 h−1) | 391 | 874 | 168 |
| Specific productivity (AU gX−1 h−1) | 195 | 192 | 139 |
| Volumetric productivity (AU L−1 h−1) | 462 | 3367 | 389 |
| Carbon-Limited Fed-Batch µset-point = 0.045 h−1 | MNLFB 3 g L−1 | ||
|---|---|---|---|
| Parameter | proROL PAOX 1 1 | proROL PGAP | proROL PAOX1 1 |
| Final activity (AU mL−1) | 147 | 341 | 358 |
| Y(P/X) (total AU total gX−1) | 1908 | 6789 | 4972 |
| Estimated µ (h−1) | 0.038 | 0.045 | 0.065 |
| qp (AU gX−1 h−1) | 68.5 | 479 | 308 |
| Specific productivity (AU gX−1 h−1) | 44 | 156 | 99 |
| Volumetric productivity (AU L−1 h−1) | 2782 | 7881 | 7160 |
| Enzyme | Expected Sequence | Actual Sequence |
|---|---|---|
| proROL | DDNLVG | EADDNL |
| proROLm | SDGGKVV | DGGKVV |
| rROL 1 | SDGGKVV | EAEFSDGGKVVAA |
| Reaction | Biocatalyst | Initial Rate (µmol Product mL−1 min−1) | Productivity (µmol Product min−1) | Half-Life (h) |
|---|---|---|---|---|
| Transesterification | EB-proROL | 27.2 | 39.43 | 498 |
| EB-rROL | 25.1 | 34.17 | 102 | |
| Esterification | EB-proROL | 308 | 4.74 | 70 |
| EB-rROL | 294 | 3.10 | 30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Fernández, J.; Benaiges, M.D.; Valero, F. Constitutive Expression in Komagataella phaffii of Mature Rhizopus oryzae Lipase Jointly with Its Truncated Prosequence Improves Production and the Biocatalyst Operational Stability. Catalysts 2021, 11, 1192. https://doi.org/10.3390/catal11101192
López-Fernández J, Benaiges MD, Valero F. Constitutive Expression in Komagataella phaffii of Mature Rhizopus oryzae Lipase Jointly with Its Truncated Prosequence Improves Production and the Biocatalyst Operational Stability. Catalysts. 2021; 11(10):1192. https://doi.org/10.3390/catal11101192
Chicago/Turabian StyleLópez-Fernández, Josu, Maria Dolors Benaiges, and Francisco Valero. 2021. "Constitutive Expression in Komagataella phaffii of Mature Rhizopus oryzae Lipase Jointly with Its Truncated Prosequence Improves Production and the Biocatalyst Operational Stability" Catalysts 11, no. 10: 1192. https://doi.org/10.3390/catal11101192