Newly Isolated Alkane Hydroxylase and Lipase Producing Geobacillus and Anoxybacillus Species Involved in Crude Oil Degradation
Abstract
:1. Introduction
2. Results
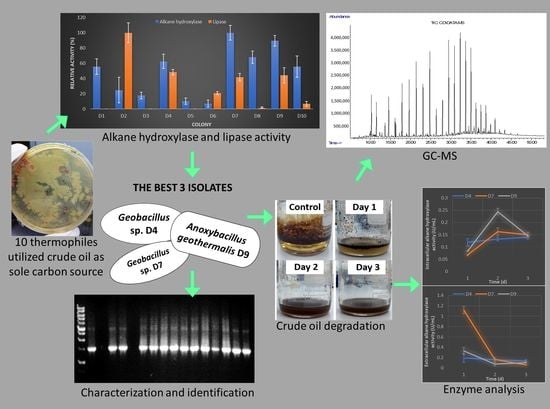
2.1. Isolation, Screening, and Selection of Thermophilic Bacteria for Crude oil Degradation
2.2. Characterization and identification of strains.
2.3. Total Protein Content and Alkane Hydroxylase for D4, D7, and D9
2.4. Gas Chromatography (GC) Analysis
3. Discussion
3.1. Growth and Enzyme Analysis of Isolates
3.2. Biodegradation of Crude Oil Compositions
4. Materials and Methods
4.1. Chemicals and Samples
4.2. Isolation and Screening of Crude Oil-Degrading Bacteria
4.3. Alkane Hydroxylase Activity
4.4. Lipase Activity
4.5. Protein Quantitation
4.6. Characterization and Identification of Strains
4.7. Construction of the Phylogenetic tree
4.8. Gas Chromatography (GC) Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chaineau, C.H.; Rougeux, G.; Yepremian, C.; Oudot, J. Effects of nutrient concentration on the biodegradation of crude oil and associated microbial populations in the soil. Soil Biol. Biochem. 2005, 37, 1490–1497. [Google Scholar] [CrossRef]
- Fernández-Álvarez, P.; Vila, J.; Garrido-Fernández, J.M.; Grifoll, M.; Lema, J.M. Trials of bioremediation on a beach affected by the heavy oil spill of the Prestige. J. Hazard. Mater. 2006, 137, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.W.G.; Chang, T.C.; Whang, L.M.; Kao, C.H.; Pan, P.T.; Cheng, S.S. Bioremediation of petroleum hydrocarbon contaminated soil: Effects of strategies and microbial community shift. Int. Biodeter. Biodegrad. 2011, 65, 1119–1127. [Google Scholar]
- Hamdoun, A.M.; Griffin, F.J.; Cherr, G.N. Tolerance to biodegraded crude oil in marine invertebrate embryos and larvae is associated with expression of a multixenobiotic resistance transporter. Aquat. Toxicol. 2002, 61, 127–140. [Google Scholar] [CrossRef]
- Zhang, Z.; Hou, Z.; Yang, C.; Ma, C.; Tao, F.; Xu, P. Degradation of n-alkanes and polycyclic aromatic hydrocarbons in petroleum by a newly isolated Pseudomonas aeruginosa DQ8. Bioresour. Technol. 2011, 102, 4111–4116. [Google Scholar] [CrossRef]
- Sood, N.; Lal, B. Isolation and characterization of a potential paraffin-wax degrading thermophilic bacterial strain Geobacillus kaustophilus TERI NSM for application in oil wells with paraffin deposition problems. Chemosphere 2008, 70, 1445–1451. [Google Scholar] [CrossRef]
- Sakthipriya, N.; Doble, M.; Sangwai, J.S. Fast degradation and viscosity reduction of waxy crude oil and model waxy crude oil using Bacillus subtilis. J. Petrol. Sci. Eng. 2015, 134, 158–166. [Google Scholar] [CrossRef]
- Xia, W.; Dong, H.; Zheng, C.; Cui, Q.; He, P.; Tang, Y. Hydrocarbon degradation by a newly isolated thermophilic Anoxybacillus sp. with bioemulsifier production and new alkB genes. RSC Adv. 2015, 5, 102367–102377. [Google Scholar] [CrossRef]
- Mulani, N.; Fulke, A.B.; D’souza, E.; Ram, A.; Maloo, A.; Sayed, F.; Gajbhiye, S.N. Biodegradation of crude oil using marine Bacillus species from Vadinar coast, Gujarat, India. Curr. Sci. 2017, 112, 569–576. [Google Scholar] [CrossRef]
- van Beilen, J.B.; Kingma, J.; Witholt, B. Substrate specificity of the alkane hydroxylase system of Pseudomonas oleovorans GPo1. Enzyme Microb. Technol. 1994, 16, 904–911. [Google Scholar] [CrossRef]
- Park, C.; Park, W. Survival and Energy Producing Strategies of Alkane Degraders under Extreme Conditions and Their Biotechnological Potential. Front. Microbiol. 2018, 9, 1081. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Miyanaga, A.; Kanaya, S.; Morikawa, M. Alkane inducible proteins in Geobacillus thermoleovorans B23. BMC Microbiol. 2009, 9, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grabowski, A.; Nercessian, O.; Fayolle, F.; Blanchet, D.; Jeanthon, C. Microbial diversity in production waters of a low-temperature biodegraded oil reservoir. FEMS Microbiol. Ecol. 2005, 54, 427–443. [Google Scholar] [CrossRef]
- Atalah, J.; Cáceres-Moreno, P.; Espina, G.; Blamey, J.M. Thermophiles and the applications of their enzymes as new biocatalysts. Bioresour. Technol. 2019, 280, 478–488. [Google Scholar] [CrossRef] [PubMed]
- Hamon, V.; Dallet, S.; Legoy, M.D. The pressure-dependence of two β-glucosidases with respect to their thermostability. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1996, 1294, 195–203. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Bao, M.; Sun, P. Metabolic pathway for a new strain Pseudomonas synxantha LSH-7′: From chemotaxis to uptake of n-hexadecane. Sci. Rep. 2017, 7, 39068. [Google Scholar] [CrossRef] [Green Version]
- Gordadze, G.N.; Poshibaeva, A.R.; Giruts, M.V.; Gayanova, A.A.; Semenova, E.M.; Koshelev, V.N. Formation of Petroleum Hydrocarbons from Prokaryote Biomass: 2. Formation of Petroleum Hydrocarbon Biomarkers from Biomass of Geobacillus jurassicus Bacteria Isolated from Crude Oil. Pet. Chem. 2018, 58, 1005–1012. [Google Scholar]
- Tourova, T.P.; Sokolova, D.S.; Semenova, E.M.; Shumkova, E.S.; Korshunova, A.V.; Babich, T.L.; Poltaraus, A.B.; Nazina, T.N. Detection of n-alkane biodegradation genes alkB and ladA in thermophilic hydrocarbon-oxidizing bacteria of the genera Aeribacillus and Geobacillus. Microbiology 2016, 85, 693–707. [Google Scholar] [CrossRef]
- Gao, Y.; Dai, J.; Peng, H.; Liu, Y.; Xu, T. Isolation and characterization of a novel organic solvent-tolerant Anoxybacillus sp. PGDY12, a thermophilic Gram-positive bacterium. J. Appl. Microbiol. 2011, 110, 472–478. [Google Scholar]
- Meng, L.; Li, W.; Bao, M.; Sun, P. Promoting the treatment of crude oil alkane pollution through the study of enzyme activity. Int. J. Biol. Macromol. 2018, 119, 708–716. [Google Scholar] [CrossRef]
- Atlas, R.M.; Bartha, R. Hydrocarbon biodegradation and oil spill bioremediation. In Advances in Microbial Ecology, 1st ed.; Marshall, K.C., Ed.; Springer: Boston, MA, USA, 1992; Volume 12, pp. 287–338. [Google Scholar]
- Gibson, D.T.; Subramanian, V.; Gibson, D.T. Microbial degradation of aromatic hydrocarbons. In Microbial Degradation of Organic Compounds, 1st ed.; Gibson, D.T., Ed.; Marcel Dekker, Inc.: New York, NY, USA, 1984; Volume 13, pp. 181–242. [Google Scholar]
- Prince, R.C.; Elmendorf, D.L.; Lute, J.R.; Hsu, C.S.; Haith, C.E.; Senius, J.D.; Butler, E.L. 17. alpha.(H)-21. beta.(H)-hopane as a conserved internal marker for estimating the biodegradation of crude oil. Environ. Sci. Technol. 1994, 28, 142–145. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.S.; Jones, D.M.; Swannell, R.P.J.; Van Duin, A.C.T. Formation of carboxylic acids during aerobic biodegradation of crude oil and evidence of microbial oxidation of hopanes. Org. Geochem. 2002, 33, 1153–1169. [Google Scholar] [CrossRef]
- Schaefer, C.E.; Yang, X.; Pelz, O.; Tsao, D.T.; Streger, S.H.; Steffan, R.J. Aerobic biodegradation of iso-butanol and ethanol and their relative effects on BTEX biodegradation in aquifer materials. Chemosphere 2010, 81, 1104–1110. [Google Scholar] [CrossRef]
- Barragan-Huerta, B.E.; Costa-Pérez, C.; Peralta-Cruz, J.; Barrera-Cortés, J.; Esparza-García, F.; Rodríguez-Vázquez, R. Biodegradation of organochlorine pesticides by bacteria grown in microniches of the porous structure of green bean coffee. Int. Biodeter. Biodegr. 2007, 59, 239–244. [Google Scholar] [CrossRef]
- Hirner, A.V.; Flassbeck, D.; Gruemping, R. Organosilicon compounds in the environment. In Oganometallic Compounds in the Environment, 2nd ed.; Peter, J.C., Ed.; John Wiley and Sons: West Sussex, UK, 2003; pp. 305–351. [Google Scholar]
- Boada, E.; Santos-Clotas, E.; Bertran, S.; Cabrera-Codony, A.; Martín, M.J.; Bañeras, L.; Gich, F. Potential use of Methylibium sp. as a biodegradation tool in organosilicon and volatile compounds removal for biogas upgrading. Chemosphere 2020, 240, 124908. [Google Scholar] [CrossRef]
- Eichhorn, E.; van der Ploeg, J.R.; Leisinger, T. Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J. Biol. Chem. 1999, 274, 26639–26646. [Google Scholar] [CrossRef] [Green Version]
- Leidner, H.; Gloor, R.; Wüest, D.; Wuhrmann, K. The influence of the sulphonic group on the biodegradability of n-alkylbenzene sulphonates. Xenobiotica 1980, 10, 47–56. [Google Scholar] [CrossRef]
- Behar, F.H.; Albrecht, P. Correlations between carboxylic acids and hydrocarbons in several crude oils. Alteration by biodegradation. Org. Geochem. 1984, 6, 597–604. [Google Scholar] [CrossRef]
- Langbehn, A.; Steinhart, H. Biodegradation studies of hydrocarbons in soils by analyzing metabolites formed. Chemosphere 1995, 30, 855–868. [Google Scholar] [CrossRef]
- Abbott, B.J.; Gledhill, W.E. The extracellular accumulation of metabolic products by hydrocarbon-degrading microorganisms. In Advances in Applied Microbiology, 1st ed.; Perlman, D., Ed.; Academic Press, Inc.: New York, NY, USA, 1971; Volume 14, pp. 249–388. [Google Scholar]
- Bushnell, L.D.; Haas, H.F. The Utilization of Certain Hydrocarbons by Microorganisms. J. Bacteriol. 1941, 41, 653–673. [Google Scholar] [CrossRef] [Green Version]
- Kester, D.R.; Duedall, I.W.; Connors, D.N.; Pytkowicz, R.M. Preparation of artificial seawater 1. Limnol. Oceanogr. 1967, 12, 176–179. [Google Scholar] [CrossRef]
- Li, P.; Wang, L.; Feng, L. Characterization of a novel Rieske-type alkane monooxygenase system in Pusillimonas sp. strain T7-7. J. Bacteriol. 2013, 195, 1892–1901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winkler, U.K.; Stuckmann, M. Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J. Bacteriol. 1979, 138, 663–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Holt, J.G.; Krieg, N.R.; Sneath, P.H.; Staley, J.T.; Williams, S.T. Bergey’s Manual of Determinative Bacteriology, 9th ed.; William & Wilkins: Baltimor, MD, USA, 1994. [Google Scholar]
- Magray, M.S.U.D.; Kumar, A.; Rawat, A.K.; Srivastava, S. Identification of Escherichia coli through analysis of 16S rRNA and 16S-23S rRNA internal transcribed spacer region sequences. Bioinformation 2011, 6, 370. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. In Mammalian Protein Metabolism, 3rd ed.; Munro, H.N., Ed.; Academic Press, Inc.: New York, NY, USA, 1969; Volume 3, pp. 22–126. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]





| Characteristics | Isolates | |||
|---|---|---|---|---|
| D4 | D7 | D9 | ||
| Colony morphology on nutrient agar | Size | Moderate | Moderate | Large |
| Pigmentation | Cream | Cream | Whitish cream | |
| Form | Circular | Circular | Circular | |
| Margin | Entire | Entire | Entire | |
| Elevation | Convex | Convex | Umbonate | |
| Shape of cells | Rod | Rod | Rod | |
| Gram staining | - | - | - | |
| Aerobic growth | + | + | + | |
| Endospores | + | + | + | |
| Catalase reaction | + | + | + | |
| Casein hydrolysis | - | - | + | |
| Starch hydrolysis | + | + | + | |
| Lipid hydrolysis | + | + | + | |
| Carbon source | Glucose | + | + | + |
| Lactose | + | + | + | |
| Sucrose | - | + | + | |
| Crude oil | + | + | + | |
| Type | RT (Min) | Compound | Molecular Formula | Degradation Percentage (%) | ||
|---|---|---|---|---|---|---|
| D4 | D7 | D9 | ||||
| n-Alkane | 5.507 | Hexane | C6H14 | 0 | –1001 | 0 |
| 9.468 | Decane | C10H22 | 100 | 100 | 100 | |
| 7.325 | Undecane | C11H24 | 40.8 | 42.4 | 51 | |
| 10.312 | Tetradecane | C14H30 | 52.8 | 63.5 | 59.1 | |
| 19.012 | Pentadecane | C15H32 | 10.9 | 12.9 | 16.9 | |
| 17.259 | Hexadecane | C16H3 | 15.8 | 21.6 | 21.4 | |
| 24.790 | Heptadecane | C17H36 | 11.5 | 20.5 | 14.3 | |
| 16.155 | Octadecane | C18H38 | 3.2 | 4.8 | –8.61 | |
| 17.908 | Nonadecane | C19H40 | 0.4 | 6.3 | 2.6 | |
| 19.661, 23.102 | Eicosane | C20H42 | 2.9 | 3.5 | 1.7 | |
| 21.414 | Heneicosane | C21H44 | 3.2 | 8.7 | 6.7 | |
| 24.141 | Docosane | C22H46 | 9.4 | 3.8 | 3.8 | |
| 27.972 | Tricosane | C23H48 | 15.3 | 26.5 | 23.3 | |
| 26.414 | Tetracosane | C24H50 | 9.5 | 16 | 13.1 | |
| 33.296 | Pentacosane | C25H52 | –0.21 | 9.0 | 7.6 | |
| 29.465 | Hexacosane | C26H54 | 14.8 | 23.5 | 15.3 | |
| 30.893 | Heptacosane | C27H56 | –1.21 | –1.81 | –2.01 | |
| 32.322 | Octacosane | C28H58 | 7.1 | 17.5 | 9.2 | |
| 33.685 | Nonacosane | C29H60 | 7.0 | 5.3 | –0.11 | |
| 34.984 | Triacontane | C30H62 | 4.5 | 7.3 | 5.8 | |
| 36.217 | Hentriacontane | C31H64 | 8.2 | 8.2 | 8.1 | |
| 37.451 | Dotriacontane | C32H66 | 7.1 | 7.5 | 2.1 | |
| 40.957 | Pentatriacontane | C35H70 | 10.9 | 7.7 | –2.01 | |
| Branched alkane | 5.442 | 3-ethylhexane | C8H18 | 31.5 | 100 | 15.5 |
| 10.052 | 2,6,10-trimethyldodecane | C15H32 | 46.0 | 52.0 | 57.3 | |
| 8.948 | 2-methyl-6-propyldodecane | C16H34 | 63.0 | 65.0 | 80.0 | |
| 13.688 | 2,6,10-trimethylpentadecane | C18H38 | 21.1 | 31.3 | 10.3 | |
| 14.597 | 2,6,10,14-tetramethylpentadecane | C19H40 | 15.6 | 17.8 | 13.6 | |
| 16.358 | 2,6,10,14-tetramethylhexadecane | C20H42 | 17.0 | 23.3 | 21.3 | |
| cylo-alkane | 18.363 | 1-Nonylcycloheptane | C16H32 | 21.0 | 37.5 | 25.2 |
| 26.738 | Cyclotetracosane | C24H48 | –6.51 | 14.5 | 11.4 | |
| 22.713 | Nonadecylcyclohexane | C25H50 | 2.7 | 9.9 | 4.0 | |
| 29.790 | 1,1,3,6-tetramethyl-2-(3,6,10,13,14-pentamethyl-3-ethyl-pentadecyl) cyclohexane | C32H64 | –4.81 | 14.2 | 14.4 | |
| Alkene | 6.871 | (2Z,4E)-3,7-Dimethyl-2,4-octadiene | C10H18 | 81.4 | 100 | 100 |
| 24.336 | 1-Tricosene | C23H46 | 0 | 0 | –1001 | |
| 32.711 | 9-Hexacosene | C26H52 | 8.4 | 10.8 | 7.3 | |
| 35.373 | 1-Hexacosene | C26H52 | 7.6 | 2.6 | 10.6 | |
| Aromatic compounds | 7.585 | 1,2,3,5-tetramethylbenzene | C10H14 | 100 | 100 | 100 |
| 10.766 | 2,3-dimethylnaphthalene | C12H12 | 48.5 | 54.6 | 54.1 | |
| 12.389 | 1,6,7-trimethylnaphthalene | C13H14 | 31.6 | 39.7 | 34.3 | |
| 20.375 | 9,10-dimethylanthracene | C16H14 | 3.2 | 20.1 | –18.71 | |
| 23.752 | 2,3,5-trimethylphenanthrene | C17H16 | 11.3 | 7.5 | 11.5 | |
| Resin | 36.802 | 28-Nor-17.beta.(H)-hopane | C29H50 | 7.0 | 10.1 | 6.2 |
| Alcohol | 6.481 | 2-Methyl-1-undecanol | C12H26O | 100 | 84.5 | 10 |
| Ester | 7.52 | 3-(Prop-2-enoyloxy) dodecane | C15H28O2 | 0 | –1001 | 0 |
| 39.074 | Octatriacontyl pentafluoropropionate | C41H77F5O2 | 7.0 | 1.5 | 6.1 | |
| Carboxylic acid | 7.975 | 2-tetradecyl ester methoxyacetic acid, | C17H34O3 | 100 | 100 | 100 |
| 9.273 | 2-oxo-methyl ester octadecanoic acid | C19H36O3 | –1001 | –13.31 | –23.41 | |
| 48.553 | 4-dodecyl dimethyl ester 1,2,4-benzenetricarboxylic acid | C23H34O6 | 0 | 0 | –1001 | |
| Organo-chlorine | 31.348 | 1-chlorooctadecane | C18H37Cl | 10.1 | 0.1 | 12.6 |
| Organo-sulfur | 8.364 | 1-octadecanesulphonyl chloride | C18H37ClO2S | 78.4 | 73.6 | 87.0 |
| 11.156 | Butyloctadecyl ester sulfurous acid | C22H46O3S | 100 | 100 | 2.4 | |
| Orgaono-silicon | 5.767 | Trichlorodocosylsilane | C22H45Cl3Si | 100 | 100 | 100 |
| 6.157 | Octamethylcyclotetrasiloxane | C8H24O4Si4 | 35.3 | 38.7 | 28.1 | |
| Biodegradation Efficiency (BE%) | 12.1 | 17.3 | 13.1 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yusoff, D.F.; Raja Abd Rahman, R.N.Z.; Masomian, M.; Ali, M.S.M.; Leow, T.C. Newly Isolated Alkane Hydroxylase and Lipase Producing Geobacillus and Anoxybacillus Species Involved in Crude Oil Degradation. Catalysts 2020, 10, 851. https://doi.org/10.3390/catal10080851
Yusoff DF, Raja Abd Rahman RNZ, Masomian M, Ali MSM, Leow TC. Newly Isolated Alkane Hydroxylase and Lipase Producing Geobacillus and Anoxybacillus Species Involved in Crude Oil Degradation. Catalysts. 2020; 10(8):851. https://doi.org/10.3390/catal10080851
Chicago/Turabian StyleYusoff, Durratul Fatini, Raja Noor Zaliha Raja Abd Rahman, Malihe Masomian, Mohd Shukuri Mohamad Ali, and Thean Chor Leow. 2020. "Newly Isolated Alkane Hydroxylase and Lipase Producing Geobacillus and Anoxybacillus Species Involved in Crude Oil Degradation" Catalysts 10, no. 8: 851. https://doi.org/10.3390/catal10080851