Synthesis of Biogenic Palladium Nanoparticles Using Citrobacter sp. for Application as Anode Electrocatalyst in a Microbial Fuel Cell
Abstract
:1. Introduction
2. Results and Discussion
2.1. Microbial Analysis
2.1.1. Culture Isolation
2.1.2. Culture Characterization
2.2. Anaerobic Pd(II) Removal Using Pure Isolates and Brits Sludge
2.2.1. Anaerobic Pd(II) Removal Using Pure Isolates
2.2.2. Comparison of Pd(II) Removal Using a Pure Isolate and Brits Sludge
2.2.3. Influence of Citrobacter sp. Live Cells on Pd(II) Removal
2.3. Abiotic Factors Influence on Pd(II) Removal by Citrobacter sp.
2.3.1. Effect of Temperature
2.3.2. Effect of pH
2.3.3. Effect of Initial Pd(II) Concentration
2.3.4. Effect of Carbon Source
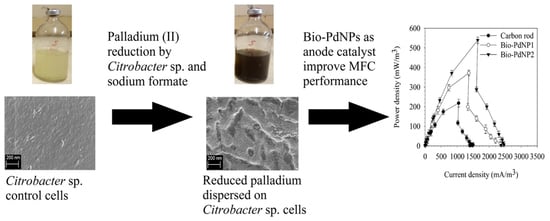
2.4. Characterization of Bio-PdNPs Synthesized by Citrobacter sp.
2.5. MFC Performance Using Bio-PdNPs Synthesized by Citrobacter sp. as Anode Catalyst
3. Materials and Methods
3.1. Microbial Culture
3.2. Microbial Isolation
3.3. Culture Characterization
3.4. Basal Mineral Media
3.5. Pd(II) Stock Solution
3.6. Anaerobic Bio-PdNPs Synthesis
3.7. Pd(II) Concentration Analysis
3.8. Bio-PdNPs Characterization
3.8.1. Morphology Analysis
3.8.2. Elemental Composition Analysis
3.8.3. XRD Analysis
3.9. Anode Preparation
3.10. MFC Set-Up and Operation
3.11. Electrochemical Analysis and Calculation
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lee, D.L.; Chang, J.S.; Lai, J.Y. Microalgae–microbial fuel cell: A mini review. Bioresour. Technol. 2015, 198, 891–895. [Google Scholar] [CrossRef] [PubMed]
- Palanisamy, G.; Jung, H.Y.; Sadhasivam, T.; Kurkuri, M.D.; Kim, S.C.; Roh, S.H. A comprehensive review on microbial fuel cell technologies: Processes, utilization, and advanced developments in electrodes and membranes. J. Clean. Prod. 2019, 221, 598–621. [Google Scholar] [CrossRef]
- Rahimnejad, M.; Adhami, A.; Darvari, S.; Zirepour, A.; Oh, S.E. Microbial fuel cell as new technology for bioelectricity generation: A review. Alex. Eng. J. 2015, 54, 745–756. [Google Scholar] [CrossRef] [Green Version]
- Chaturvedi, V.; Verma, P. Microbial fuel cell: A green approach for the utilization of waste for the generation of bioelectricity. Bioresour. Bioprocess. 2016, 3, 38. [Google Scholar] [CrossRef] [Green Version]
- Osman, M.H.; Shah, A.A.; Walsh, F.C. Recent progress and continuing challenges in bio-fuel cells. part II: Microbial. Biosens. Bioelectron. 2010, 26, 953–963. [Google Scholar] [CrossRef] [PubMed]
- Quan, X.; Sun, B.; Xu, H. Anode decoration with biogenic Pd nanoparticles improved power generation in microbial fuel cells. Electrochim. Acta 2015, 182, 815–820. [Google Scholar] [CrossRef]
- Cai, H.; Wang, J.; Bu, Y.; Zhong, Q. Treatment of carbon cloth anodes for improving power generation in a dual-chamber microbial fuel cell. J. Chem. Technol. Biotechnol. 2013, 88, 623–628. [Google Scholar] [CrossRef]
- Zhu, N.; Chen, X.; Zhang, T.; Wu, P.; Li, P.; Wu, J. Improved performance of membrane free single-chamber air-cathode microbial fuel cells with nitric acid and ethylenediamine surface modified activated carbon fiber felt anodes. Bioresour. Technol. 2011, 102, 422–426. [Google Scholar] [CrossRef]
- Sun, M.; Zhang, F.; Tong, Z.H.; Sheng, G.P.; Chen, Y.Z.; Zhao, Y.; Chen, Y.P.; Zhou, S.Y.; Liu, G.; Tian, Y.C.; et al. A gold-sputtered carbon paper as an anode for improved electricity generation from a microbial fuel cell inoculated with Shewanella oneidensis MR-1. Biosens. Bioelectron. 2010, 26, 338–343. [Google Scholar] [CrossRef]
- Fan, Y.; Xu, S.; Schaller, R.; Jiao, J.; Chaplen, F.; Liu, H. Nanoparticle decorated anodes for enhanced current generation in microbial electrochemical cells. Biosens. Bioelectron. 2011, 26, 1908–1912. [Google Scholar] [CrossRef]
- Kumar, R.; Singh, L.; Zularisam, A.W. Exoelectrogens: Recent advances in molecular drivers involved in extracellular electron transfer and strategies used to improve it for microbial fuel cell applications. Renew. Sustain. Energy Rev. 2016, 56, 1322–1336. [Google Scholar] [CrossRef] [Green Version]
- Rosenbaum, M.; Schröder, U.; Scholz, F. Investigation of the electrocatalytic oxidation of formate and ethanol at platinum black under microbial fuel cell conditions. J. Solid State Electrochem. 2006, 10, 872–878. [Google Scholar] [CrossRef]
- Varanasi, J.L.; Nayak, A.K.; Sohn, Y.; Pradhan, D.; Das, D. Improvement of power generation of microbial fuel cell by integrating tungsten oxide electrocatalyst with pure or mixed culture biocatalysts. Electrochim. Acta 2016, 199, 154–163. [Google Scholar] [CrossRef]
- Gupta, R.; Guin, S.K.; Aggarwal, S.K. Electrocrystallization of palladium (Pd) nanoparticles on platinum (Pt) electrode and its application for electro-oxidation of formic acid and methanol. Electrochim. Acta 2014, 116, 314–320. [Google Scholar] [CrossRef]
- Zhou, Z.H.; Li, W.S.; Fu, Z.; Xiang, X.D. Carbon nanotube-supported Pt-HxMoO3 as electrocatalyst for methanol oxidation. Int. J. Hydrogen Energy 2010, 35, 936–941. [Google Scholar] [CrossRef]
- Quan, X.; Zhang, X.; Sun, Y.; Zhao, J. Iohexol degradation by biogenic palladium nanoparticles hosted in anaerobic granular sludge. Front. Microbiol. 2018, 9, 1980. [Google Scholar] [CrossRef]
- Yang, G.; Chen, D.; Lv, P.; Kong, X.; Sun, Y.; Wang, Z.; Yuan, Z.; Liu, H.; Yang, J. Core-shell Au-Pd nanoparticles as cathode catalysts for microbial fuel cell applications. Sci. Rep. 2016, 6, 35252. [Google Scholar] [CrossRef] [Green Version]
- Moon, J.S.; Lee, Y.W.; Han, S.B.; Park, K.W. Pd nanoparticles on mesoporous tungsten carbide as a non-Pt electrocatalyst for methanol electrooxidation reaction in alkaline solution. Int. J. Hydrogen Energy 2014, 39, 7798–7804. [Google Scholar] [CrossRef]
- Hennebel, T.; Verhagen, P.; Simoen, H.; Gusseme, B.D.; Vlaeminck, S.E.; Boon, N.; Verstraete, W. Remediation of trichloroethylene by bio-precipitated and encapsulated palladium nanoparticles in a fixed bed reactor. Chemosphere 2009, 76, 1221–1225. [Google Scholar] [CrossRef]
- Baxter-Plant, V.S.; Mikheenko, I.P.; Macaskie, L.E. Sulphate-reducing bacteria, palladium and the reductive dehalogenation of chlorinated aromatic compounds. Biodegradation 2003, 14, 83–90. [Google Scholar] [CrossRef]
- Malunga, K.; Chirwa, E. Redox potential and proton demand in an anaerobic palladium (II) reducing culture of Desulfovibrio desulfuricans Seroval. Chem. Eng. Trans. 2019, 76, 1309–1314. [Google Scholar] [CrossRef]
- Yates, M.D.; Logan, B.E. Biotemplated palladium catalysts can be stabilized on different support materials. ChemElectroChem 2014, 1, 1867–1873. [Google Scholar] [CrossRef]
- Han, R.; Li, P.; Liu, T.; Li, X.; Wu, Y.; Wang, Y.; Chen, D. Effects of incubation conditions on Cr(VI) reduction by c-type cytochromes in intact Shewanella oneidensis MR-1 cells. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Sharmila, G.; Farzana Fathima, M.; Haries, S.; Geetha, S.; Manoj Kumar, N.; Muthukumaran, C. Green synthesis, characterization and antibacterial efficacy of palladium nanoparticles synthesized using Filicium decipiens leaf extract. J. Mol. Struct. 2017, 1138, 35–40. [Google Scholar] [CrossRef]
- Adams, C.P.; Walker, K.A.; Obare, S.O.; Docherty, K.M. Size-dependent antimicrobial effects of novel palladium nanoparticles. PLoS ONE 2014, 9, e85981. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.Z.; Lee, S.D.; Bhatnagar, R.S. Toxicity of palladium. Toxicol. Lett. 1979, 4, 469–473. [Google Scholar] [CrossRef]
- Yong, P.; Rowson, N.A.; Farr, J.P.G.; Harris, I.R.; Macaskie, L.E. Bioreduction and biocrystallization of palladium by Desulfovibrio desulfuricans NCIMB 8307. Biotechnol. Bioeng. 2002, 80, 369–379. [Google Scholar] [CrossRef]
- Hosseinkhani, B.; Hennebel, T.; Van Nevel, S.; Verschuere, S.; Yakimov, M.M.; Cappello, S.; Blaghen, M.; Boon, N. Biogenic nanopalladium based remediation of chlorinated hydrocarbons in marine environments. Environ. Sci. Technol. 2014, 48, 550–557. [Google Scholar] [CrossRef]
- Hennebel, T.; Van Nevel, S.; Verschuere, S.; De Corte, S.; De Gusseme, B.; Cuvelier, C.; Fitts, J.P.; van der Lelie, D.; Boon, N.; Verstraete, W. Palladium nanoparticles produced by fermentatively cultivated bacteria as catalyst for diatrizoate removal with biogenic hydrogen. Appl. Microbiol. Biotechnol. 2011, 91, 1435–1445. [Google Scholar] [CrossRef]
- Wang, J.; Bi, S.; Chen, Y.; Hu, Y. Electron transfer involved in bio-Pd (0) synthesis by Citrobacter freundii at different growth phases. Ecotoxicol. Environ. Saf. 2020, 190, 110124. [Google Scholar] [CrossRef]
- Hobman, J.L.; Crossman, L.C. Bacterial antimicrobial metal ion resistance. J. Med. Microbiol. 2015, 64, 471–497. [Google Scholar] [CrossRef] [PubMed]
- Molina, M.d.C.; Bautista, L.F.; Belda, I.; Carmona, M.; Díaz, E.; Durante-Rodríguez, G.; García-Salgado, S.; Jaime, L.-A.; Martínez-Hidalgo, P.; Quijano, M.Á.; et al. Bioremediation of soil contaminated with arsenic. In Microbes and Enzymes in Soil Health and Bioremediation; Kumar, A., Sharma, S., Eds.; Springer: Singapore, 2019; pp. 321–351. [Google Scholar]
- Sannasi, P.; Kader, J.; Ismail, B.S.; Salmijah, S. Sorption of Cr (VI), Cu (II) and Pb (II) by growing and non-growing cells of a bacterial consortium. Bioresour. Technol. 2006, 97, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Anna, M.; Renata, M.; Jana, K.; Oksana, V.; Magdalena, B. Influence of used bacterial culture on zinc and aluminium bioleaching from printed circuit boards. Nova Biotechnol. Chim. 2015, 14, 45–51. [Google Scholar] [CrossRef] [Green Version]
- Simona, K.; Ľudmila, H.; Peter, P. Zinc bioaccumulation by microbial consortium isolated from nickel smelter sludge disposal site. Nova Biotechnol. Chim. 2017, 16, 48–53. [Google Scholar] [CrossRef] [Green Version]
- de Vargas, I.; Macaskie, L.E.; Guibal, E. Biosorption of palladium and platinum by sulfate-reducing bacteria. J. Chem. Technol. Biotechnol. 2004, 79, 49–56. [Google Scholar] [CrossRef]
- Windt, W.D.; Aelterman, P.; Verstraete, W. Bioreductive deposition of palladium (0) nanoparticles on Shewanella oneidensis with catalytic activity towards reductive dechlorination of polychlorinated biphenyls. Environ. Microbiol. 2005, 7, 314–325. [Google Scholar] [CrossRef] [PubMed]
- Bunge, M.; Søbjerg, L.S.; Rotaru, A.-E.; Gauthier, D.; Lindhardt, A.T.; Hause, G.; Finster, K.; Kingshott, P.; Skrydstrup, T.; Meyer, R.L. Formation of palladium (0) nanoparticles at microbial surfaces. Biotechnol. Bioeng. 2010, 107, 206–215. [Google Scholar] [CrossRef]
- Deplanche, K.; Caldelari, I.; Mikheenko, I.P.; Sargent, F.; Macaskie, L.E. Involvement of hydrogenases in the formation of highly catalytic Pd (0) nanoparticles by bioreduction of Pd (II) using Escherichia coli mutant strains. Microbiology 2010, 156, 2630–2640. [Google Scholar] [CrossRef] [Green Version]
- Camargo, F.A.O.; Bento, F.M.; Okeke, B.C.; Frankenberger, W.T. Chromate reduction by chromium-resistant bacteria isolated from soils contaminated with dichromate. J. Environ. Qual. 2003, 32, 1228–1233. [Google Scholar] [CrossRef]
- Robinson, P.K. Enzymes: Principles and biotechnological applications. Essays Biochem. 2015, 59, 1–41. [Google Scholar] [CrossRef]
- White, C.P.; Popovici, J.; Lytle, D.A.; Adcock, N.J.; Rice, E.W. Effect of pH on the electrophoretic mobility of spores of bacillus anthracis and its surrogates in aqueous solutions. Appl. Environ. Microbiol. 2012, 78, 8470–8473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Remoudaki, E.; Tsezos, M.; Hatzikioseyian, A.; Karakoussis, V. Mechanism of palladium biosorption by microbial biomass. The effects of metal ionic speciation and solution co-ions. In Process Metallurgy; Amils, R., Ballester, A., Eds.; Elsevier: Amsterdam, The Netherlands, 1999; Volume 9, pp. 449–462. [Google Scholar]
- Jucker, B.A.; Harms, H.; Zehnder, A.J. Adhesion of the positively charged bacterium Stenotrophomonas (Xanthomonas) maltophilia 70401 to glass and Teflon. J. Bacteriol. 1996, 178, 5472–5479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, S.A. Experimental determination of the hydrolysis constants of Pt2+ and Pd2+ at 25°C from the solubility of Pt and Pd in aqueous hydroxide solutions. Geochim. Cosmochim. Acta 1991, 55, 1759–1767. [Google Scholar] [CrossRef]
- Kielhorn, J.; Melber, C.; Keller, D.; Mangelsdorf, I. Palladium—A review of exposure and effects to human health. Int. J. Hyg. Environ. Health 2002, 205, 417–432. [Google Scholar] [CrossRef] [PubMed]
- Crable, B.R.; Plugge, C.M.; McInerney, M.J.; Stams, A.J.M. Formate formation and formate conversion in biological fuels production. Enzyme Res. 2011, 2011, 532536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hazarika, M.; Borah, D.; Bora, P.; Silva, A.R.; Das, P. Biogenic synthesis of palladium nanoparticles and their applications as catalyst and antimicrobial agent. PLoS ONE 2017, 12, e0184936. [Google Scholar] [CrossRef]
- Chae, K.J.; Choi, M.J.; Lee, J.W.; Kim, K.Y.; Kim, I.S. Effect of different substrates on the performance, bacterial diversity, and bacterial viability in microbial fuel cells. Bioresour. Technol. 2009, 100, 3518–3525. [Google Scholar] [CrossRef]
- Quan, X.; Xu, H.; Sun, B.; Xiao, Z. Anode modification with palladium nanoparticles enhanced Evans Blue removal and power generation in microbial fuel cells. Int. Biodeterior. Biodegrad. 2018, 132, 94–101. [Google Scholar] [CrossRef]
- Ali, J.; Sohail, A.; Wang, L.; Rizwan Haider, M.; Mulk, S.; Pan, G. Electro-microbiology as a promising approach towards renewable energy and environmental sustainability. Energies 2018, 11, 1822. [Google Scholar] [CrossRef] [Green Version]
- Mtimunye, P.J. A Steady-State Model for Hexavalent Chromium Reduction in Simulated Biological Reactive Barrier: Microcosm Analysis. Ph.D. Thesis, University of Pretoria, Pretoria, South Africa, 2011. [Google Scholar]
- Wang, W.; Zhang, B.; Liu, Q.; Du, P.; Liu, W.; He, Z. Biosynthesis of palladium nanoparticles using Shewanella loihica PV-4 for excellent catalytic reduction of chromium (vi). Environ. Sci. Nano 2018, 5, 730–739. [Google Scholar] [CrossRef]
- Rava, E.M.E.; Chirwa, E.M.N. Effect of carrier fill ratio on biofilm properties and performance of a hybrid fixed-film bioreactor treating coal gasification wastewater for the removal of COD, phenols and ammonia-nitrogen. Water Sci. Technol. 2016, 73, 2461–2467. [Google Scholar] [CrossRef] [PubMed]










| Species Identified | Query Cover % |
|---|---|
| Exigobacterium sp. | 100 |
| Bacillus sp. | 100 |
| Citrobacter sp. | 100 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matsena, M.T.; Tichapondwa, S.M.; Chirwa, E.M.N. Synthesis of Biogenic Palladium Nanoparticles Using Citrobacter sp. for Application as Anode Electrocatalyst in a Microbial Fuel Cell. Catalysts 2020, 10, 838. https://doi.org/10.3390/catal10080838
Matsena MT, Tichapondwa SM, Chirwa EMN. Synthesis of Biogenic Palladium Nanoparticles Using Citrobacter sp. for Application as Anode Electrocatalyst in a Microbial Fuel Cell. Catalysts. 2020; 10(8):838. https://doi.org/10.3390/catal10080838
Chicago/Turabian StyleMatsena, Mpumelelo Thomas, Shepherd Masimba Tichapondwa, and Evans Martin Nkhalambayausi Chirwa. 2020. "Synthesis of Biogenic Palladium Nanoparticles Using Citrobacter sp. for Application as Anode Electrocatalyst in a Microbial Fuel Cell" Catalysts 10, no. 8: 838. https://doi.org/10.3390/catal10080838