Comparing Genetic Risk and Clinical Risk Classification in Luminal-like Breast Cancer Patients Using a 23-Gene Classifier
Abstract
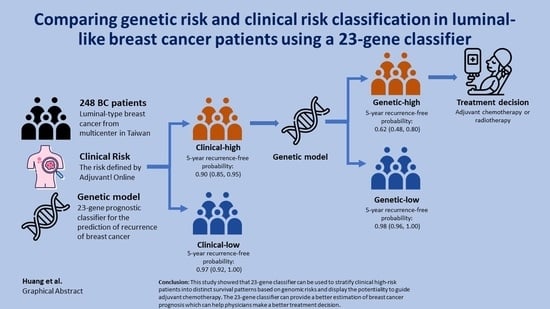
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. The 23-Gene Classifier
2.3. Prognostic and Statistical Analysis
3. Results
3.1. Baseline Demography of Enrolled Population
3.2. The Clinical Performance between Two Prognosis Tools
3.3. RFI Analysis and Cox Proportional Hazards Regression Model
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ER | estrogen receptor |
| FFPE | formalin-fixed paraffin-embedded |
| HER2 | human epidermal growth factor receptor 2 |
| HR | hormone receptor |
| IHC | immunohistochemistry |
| LVI | lymphovascular invasion |
| MGA | multi-gene expression assays |
| PR | progesterone receptor |
| RFI | recurrence-free interval |
| ROC | receiver operating characteristic |
| RT-PCR | reverse transcriptase-polymerase chain reaction |
References
- Health Promotion Administration Ministry of Health; WelFare, T. Cancer Registry Annual Report, 2018 Taiwan; Health Promotion Administration Ministry of Health: Taiwan, China, 2020. [Google Scholar]
- Yeo, B.; Zabaglo, L.; Hills, M.; Dodson, A.; Smith, I.; Dowsett, M. Clinical utility of the IHC4+C score in oestrogen receptor-positive early breast cancer: A prospective decision impact study. Br. J. Cancer 2015, 113, 390–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheang, M.C.U.; Bliss, J.M.; Viale, G.; Speirs, V.; Palmieri, C.; Shaaban, A.; Lønning, P.E.; Morden, J.; Porta, N.; Jassem, J.; et al. Evaluation of applying IHC4 as a prognostic model in the translational study of Intergroup Exemestane Study (IES): PathIES. Breast Cancer Res. Treat. 2017, 168, 169–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burguin, A.; Diorio, C.; Durocher, F. Breast Cancer Treatments: Updates and New Challenges. J. Pers. Med. 2021, 11, 808. [Google Scholar] [CrossRef] [PubMed]
- Early Breast Cancer Trialists’ Collaborative Group. Tamoxifen for early breast cancer: An overview of the randomised trials. Lancet 1998, 351, 1451–1467. [Google Scholar] [CrossRef]
- Guler, E.N. Gene Expression Profiling in Breast Cancer and Its Effect on Therapy Selection in Early-Stage Breast Cancer. Eur. J. Breast Health 2017, 13, 168–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- NCCN. National Comprehensive Cancer Network. In Breast Cancer (Version 7.2021); NCCN: Plymouth Meeting, PA, USA, 2021. [Google Scholar]
- Vieira, A.; Schmitt, F. An Update on Breast Cancer Multigene Prognostic Tests-Emergent Clinical Biomarkers. Front. Med. 2018, 5, 248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.-H.; Wei, J.-R.; Lei, J.; Chiu, J.-Y.; Shih, K.-H. A Clinicogenetic Prognostic Classifier for Prediction of Recurrence and Survival in Asian Breast Cancer Patients. Front. Oncol. 2021, 11, 645853. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, F.; van’t Veer, L.J.; Bogaerts, J.; Slaets, L.; Viale, G.; Delaloge, S.; Pierga, J.Y.; Brain, E.; Causeret, S.; Delorenzi, M.; et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N. Engl. J. Med. 2016, 375, 717–729. [Google Scholar] [CrossRef] [Green Version]
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E., Jr.; Dees, E.C.; Goetz, M.P.; Olson, J.A.; et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med. 2018, 379, 111–121. [Google Scholar] [CrossRef] [Green Version]
- Arango, B.A.; Rivera, C.L.; Gluck, S. Gene expression profiling in breast cancer. Am. J. Transl. Res. 2013, 5, 132–138. [Google Scholar] [PubMed]
- Bueno-De-Mesquita, J.M.; van Harten, W.H.; Retel, V.P.; Veer, L.J.V.; van Dam, F.S.; Karsenberg, K.; Douma, K.F.; van Tinteren, H.; Peterse, J.L.; Wesseling, J.; et al. Use of 70-gene signature to predict prognosis of patients with node-negative breast cancer: A prospective community-based feasibility study (RASTER). Lancet Oncol. 2007, 8, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-H.; Chiu, J.-Y.; Shih, K.-H. A 23-gene prognostic classifier for prediction of recurrence and survival for Asian breast cancer patients. Biosci. Rep. 2020, 40, BSR20202794. [Google Scholar] [CrossRef] [PubMed]
- Barry, W.T.; Kernagis, D.N.; Dressman, H.K.; Griffis, R.J.; Hunter, J.D.; Olson, J.A.; Marks, J.R.; Ginsburg, G.S.; Marcom, P.K.; Nevins, J.R.; et al. Intratumor heterogeneity and precision of microarray-based predictors of breast cancer biology and clinical outcome. J. Clin. Oncol. 2010, 28, 2198–2206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filipits, M.; Rudas, M.; Jakesz, R.; Dubsky, P.; Fitzal, F.; Singer, C.F.; Dietze, O.; Greil, R.; Jelen, A.; Sevelda, P.; et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin. Cancer Res. 2011, 17, 6012–6020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, J.S.; Mullins, M.; Cheang, M.C.U.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Buus, R.; Sestak, I.; Kronenwett, R.; Denkert, C.; Dubsky, P.; Krappmann, K.; Scheer, M.; Petry, C.; Cuzick, J.; Dowsett, M. Comparison of EndoPredict and EPclin With Oncotype DX Recurrence Score for Prediction of Risk of Distant Recurrence After Endocrine Therapy. J. Natl. Cancer Inst. 2016, 108, djw149. [Google Scholar] [CrossRef] [PubMed]
- Lænkholm, A.V.; Jensen, M.B.; Eriksen, J.O.; Rasmussen, B.B.; Knoop, A.S.; Buckingham, W.; Ferree, S.; Schaper, C.; Nielsen, T.O.; Haffner, T.; et al. PAM50 Risk of Recurrence Score Predicts 10-Year Distant Recurrence in a Comprehensive Danish Cohort of Postmenopausal Women Allocated to 5 Years of Endocrine Therapy for Hormone Receptor-Positive Early Breast Cancer. J. Clin. Oncol. 2018, 36, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Kalinsky, K.; Barlow, W.E.; Meric-Bernstam, F.; Gralow, J.R.; Albain, K.S.; Hayes, D.; Lin, N.; Perez, E.A.; Goldstein, L.J.; Chia, S.; et al. Abstract GS3-00: First results from a phase III randomized clinical trial of standard adjuvant endocrine therapy (ET) +/- chemotherapy (CT) in patients (pts) with 1-3 positive nodes, hormone receptor-positive (HR+) and HER2-negative (HER2-) breast cancer (BC) with recurrence score (RS) < 25: SWOG S1007 (RxPonder). Cancer Res. 2021, 81 (Suppl. S4), GS3-00. [Google Scholar]
- Lei, L.; Chan, H.-C.; Jia, W.X.; Lu, T.-P.; Cheng, S.H.-C. Recurrence risk stratification by Dutch clinical risk criteria for early-stage luminal breast cancer patients in Taiwan: A population-based analysis. J. Clin. Oncol. 2020, 38 (Suppl. S15), e12509. [Google Scholar] [CrossRef]
- Schreuder, K.; Kuijer, A.; Bentum, S.; van Dalen, T.; Siesling, S. Use and Impact of the 21-Gene Recurrence Score in Relation to the Clinical Risk of Developing Metastases in Early Breast Cancer Patients in the Netherlands. Public Health Genom. 2018, 21, 85–92. [Google Scholar] [CrossRef] [PubMed]








| Characteristic | Overall | Clinical Risk | ||
|---|---|---|---|---|
| N = 248 1 | Low, N = 69 1 | High, N = 179 1 | p-Value 2 | |
| Age | 0.8 | |||
| 40–60 | 168 (67.74%) | 49 (71.01%) | 119 (66.48%) | |
| >60 | 62 (25.00%) | 16 (23.19%) | 46 (25.70%) | |
| <40 | 18 (7.26%) | 4 (5.80%) | 14 (7.82%) | |
| Tumor stage | <0.001 | |||
| T1 | 114 (45.97%) | 63 (91.30%) | 51 (28.49%) | |
| T2 | 123 (49.60%) | 6 (8.70%) | 117 (65.36%) | |
| T3 | 11 (4.44%) | 0 (0.00%) | 11 (6.15%) | |
| N stage | <0.001 | |||
| N0 | 178 (71.77%) | 65 (94.20%) | 113 (63.13%) | |
| N1 | 59 (23.79%) | 4 (5.80%) | 55 (30.73%) | |
| N2 | 11 (4.44%) | 0 (0.00%) | 11 (6.15%) | |
| LVI | <0.001 | |||
| No | 190 (76.61%) | 66 (95.65%) | 124 (69.27%) | |
| Yes | 58 (23.39%) | 3 (4.35%) | 55 (30.73%) | |
| Grade | <0.001 | |||
| I | 95 (38.31%) | 21 (30.43%) | 74 (41.34%) | |
| II | 130 (52.42%) | 48 (69.57%) | 82 (45.81%) | |
| III | 23 (9.27%) | 0 (0.00%) | 23 (12.85%) | |
| Chemotherapy | <0.001 | |||
| No | 156 (62.90%) | 55 (79.71%) | 101 (56.42%) | |
| Yes | 92 (37.10%) | 14 (20.29%) | 78 (43.58%) | |
| Radiotherapy | 0.10 | |||
| No | 145 (58.47%) | 46 (66.67%) | 99 (55.31%) | |
| Yes | 103 (41.53%) | 23 (33.33%) | 80 (44.69%) | |
| Hormonal therapy | 0.11 | |||
| No | 8 (3.23%) | 0 (0.00%) | 8 (4.47%) | |
| Yes | 240 (96.77%) | 69 (100.0%) | 171 (95.53%) | |
| Relapse | 0.11 | |||
| No | 221 (89.11%) | 65 (94.20%) | 156 (87.15%) | |
| Yes | 27 (10.89%) | 4 (5.80%) | 23 (12.85%) | |
| 23-gene classifier | 0.2 | |||
| Low | 195 (78.63%) | 58 (84.06%) | 137 (76.54%) | |
| High | 53 (21.37%) | 11 (15.94%) | 42 (23.46%) | |
| Follow-up | 67.70 [43.33, 97.55] | 62.23 [36.26, 102.77] | 68.46 [45.31, 92.70] | 0.8 |
| Chemotherapy | |||
|---|---|---|---|
| Characteristic | No, N = 156 1 | Yes, N = 92 1 | p-Value 2 |
| Age | 0.3 | ||
| 40–60 | 101 (64.74%) | 67 (72.83%) | |
| >60 | 44 (28.21%) | 18 (19.57%) | |
| <40 | 11 (7.05%) | 7 (7.61%) | |
| Tumor stage | <0.001 | ||
| T1 | 83 (53.21%) | 31 (33.70%) | |
| T2 | 71 (45.51%) | 52 (56.52%) | |
| T3 | 2 (1.28%) | 9 (9.78%) | |
| N stage | <0.001 | ||
| N0 | 129 (82.69%) | 49 (53.26%) | |
| N1 | 25 (16.03%) | 34 (36.96%) | |
| N2 | 2 (1.28%) | 9 (9.78%) | |
| LVI | 0.044 | ||
| No | 126 (80.77%) | 64 (69.57%) | |
| Yes | 30 (19.23%) | 28 (30.43%) | |
| Grade | 0.008 | ||
| I | 66 (42.31%) | 29 (31.52%) | |
| II | 82 (52.56%) | 48 (52.17%) | |
| III | 8 (5.13%) | 15 (16.30%) | |
| Relapse | 0.012 | ||
| No | 145 (92.95%) | 76 (82.61%) | |
| Yes | 11 (7.05%) | 16 (17.39%) | |
| Clinical risk | <0.001 | ||
| Low | 55 (35.26%) | 14 (15.22%) | |
| High | 101 (64.74%) | 78 (84.78%) | |
| 23-gene classifier | 0.3 | ||
| Low | 126 (80.77%) | 69 (75.00%) | |
| High | 30 (19.23%) | 23 (25.00%) | |
| Follow-up | 67.03 [40.64, 99.79] | 68.43 [44.78, 89.02] | 0.8 |
| Radiotherapy | |||
|---|---|---|---|
| Characteristic | No, N = 145 1 | Yes, N = 103 1 | p-Value 2 |
| Age | 0.076 | ||
| 40–60 | 96 (66.21%) | 72 (69.90%) | |
| >60 | 42 (28.97%) | 20 (19.42%) | |
| <40 | 7 (4.83%) | 11 (10.68%) | |
| Tumor stage | 0.024 | ||
| T1 | 68 (46.90%) | 46 (44.66%) | |
| T2 | 75 (51.72%) | 48 (46.60%) | |
| T3 | 2 (1.38%) | 9 (8.74%) | |
| N stage | 0.6 | ||
| N0 | 104 (71.72%) | 74 (71.84%) | |
| N1 | 36 (24.83%) | 23 (22.33%) | |
| N2 | 5 (3.45%) | 6 (5.83%) | |
| LVI | >0.9 | ||
| No | 111 (76.55%) | 79 (76.70%) | |
| Yes | 34 (23.45%) | 24 (23.30%) | |
| Grade | 0.2 | ||
| I | 51 (35.17%) | 44 (42.72%) | |
| II | 83 (57.24%) | 47 (45.63%) | |
| III | 11 (7.59%) | 12 (11.65%) | |
| Relapse | 0.2 | ||
| No | 132 (91.03%) | 89 (86.41%) | |
| Yes | 13 (8.97%) | 14 (13.59%) | |
| Clinical risk | 0.10 | ||
| Low | 46 (31.72%) | 23 (22.33%) | |
| High | 99 (68.28%) | 80 (77.67%) | |
| 23-gene classifier | 0.12 | ||
| Low | 109 (75.17%) | 86 (83.50%) | |
| High | 36 (24.83%) | 17 (16.50%) | |
| Follow-up | 77.01 [48.20, 102.77] | 56.75 [30.90, 76.11] | <0.001 |
| Characteristic | Clinical Outcome: Relapse | Total | |
|---|---|---|---|
| Yes | No | ||
| 23-gene classifier (high/low) | |||
| High | 22 | 5 | 27 |
| Low | 31 | 190 | 221 |
| Total | 53 | 195 | 248 |
| Characteristic | Clinical Outcome: Relapse | Total | |
|---|---|---|---|
| Yes | No | ||
| Clinical risk (high/low) | |||
| High | 23 | 4 | 27 |
| Low | 156 | 65 | 221 |
| Total | 179 | 69 | 248 |
| Characteristic | Univariate Analysis | Model 1 | Model 2 | Model 3 | Model 4 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR 1 | 95% CI 1 | p-Value | HR 1 | 95% CI 1 | p-Value | HR 1 | 95% CI 1 | p-Value | HR 1 | 95% CI 1 | p-Value | HR 1 | 95% CI 1 | p-Value | |
| Age | |||||||||||||||
| 40–60 | — | — | — | — | — | — | — | — | — | — | |||||
| >60 | 1.31 | 0.45, 3.82 | 0.625 | 1.46 | 0.49, 4.36 | 0.5 | 1.25 | 0.42, 3.69 | 0.7 | 1.45 | 0.48, 4.33 | 0.5 | 1.47 | 0.49, 4.42 | 0.5 |
| <40 | 2.96 | 0.81, 10.7 | 0.100 | 2.50 | 0.59, 10.5 | 0.2 | 3.31 | 0.84, 13.1 | 0.087 | 2.39 | 0.56, 10.2 | 0.2 | 2.31 | 0.54, 9.86 | 0.3 |
| LVI | |||||||||||||||
| No | — | — | — | — | — | — | — | — | — | — | |||||
| Yes | 0.69 | 0.20, 2.38 | 0.555 | 0.37 | 0.09, 1.56 | 0.2 | 0.17 | 0.04, 0.68 | 0.012 | 0.37 | 0.09, 1.55 | 0.2 | 0.38 | 0.09, 1.62 | 0.2 |
| N stage | |||||||||||||||
| N0 | — | — | — | — | — | — | — | — | — | — | |||||
| N1 | 4.08 | 1.37, 12.1 | 0.012 | 3.24 | 0.67, 15.6 | 0.14 | 6.55 | 1.62, 26.5 | 0.008 | 2.89 | 0.54, 15.5 | 0.2 | 2.73 | 0.51, 14.5 | 0.2 |
| N2 | 15.8 | 4.82, 51.8 | <0.001 | 7.65 | 1.40, 41.8 | 0.019 | 26.6 | 5.65, 126 | <0.001 | 6.77 | 1.12, 40.9 | 0.037 | 6.25 | 1.03, 38.0 | 0.047 |
| Tumor grade | |||||||||||||||
| Grade I | — | — | — | — | — | — | — | — | — | — | |||||
| Grade II | 3.54 | 1.00, 12.5 | 0.050 | 1.40 | 0.35, 5.55 | 0.6 | 2.40 | 0.65, 8.92 | 0.2 | 1.43 | 0.36, 5.77 | 0.6 | 1.38 | 0.34, 5.58 | 0.6 |
| Grade III | 5.18 | 1.04, 25.7 | 0.044 | 2.00 | 0.34, 11.8 | 0.4 | 5.10 | 0.89, 29.0 | 0.067 | 1.91 | 0.32, 11.3 | 0.5 | 1.78 | 0.30, 10.7 | 0.5 |
| Chemotherapy | |||||||||||||||
| No | — | — | — | — | — | — | — | — | — | — | |||||
| Yes | 2.08 | 0.82, 5.27 | 0.122 | 0.69 | 0.19, 2.52 | 0.6 | 0.63 | 0.20, 1.98 | 0.4 | 0.69 | 0.19, 2.48 | 0.6 | 0.69 | 0.19, 2.46 | 0.6 |
| 23-gene classifier | |||||||||||||||
| Low | — | — | — | — | — | — | — | — | |||||||
| High | 20.9 | 6.04, 72.1 | <0.001 | 10.5 | 2.65, 41.8 | <0.001 | 10.5 | 2.63, 42.2 | <0.001 | 5.60 | 0.35, 90.8 | 0.2 | |||
| Clinical risk | |||||||||||||||
| Low | — | — | — | — | — | — | — | — | |||||||
| High | 2.92 | 0.67, 12.7 | 0.153 | 1.59 | 0.30, 8.35 | 0.6 | 1.38 | 0.25, 7.80 | 0.7 | 0.85 | 0.07, 9.92 | 0.9 | |||
| Interaction of clinical risk and 23-gene classifier | |||||||||||||||
| High * High | 2.33 | 0.09, 62.2 | 0.6 | ||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, C.-C.; Chen, T.-H.; Liu, L.-C.; Huang, C.-S.; Liang, J.-A.; Hsu, Y.-C.; Hsieh, C.-M.; Huang, S.-L.; Shih, K.-H.; Tseng, L.-M. Comparing Genetic Risk and Clinical Risk Classification in Luminal-like Breast Cancer Patients Using a 23-Gene Classifier. Cancers 2022, 14, 6263. https://doi.org/10.3390/cancers14246263
Huang C-C, Chen T-H, Liu L-C, Huang C-S, Liang J-A, Hsu Y-C, Hsieh C-M, Huang S-L, Shih K-H, Tseng L-M. Comparing Genetic Risk and Clinical Risk Classification in Luminal-like Breast Cancer Patients Using a 23-Gene Classifier. Cancers. 2022; 14(24):6263. https://doi.org/10.3390/cancers14246263
Chicago/Turabian StyleHuang, Chi-Cheng, Ting-Hao Chen, Liang-Chih Liu, Chiun-Sheng Huang, Ji-An Liang, Yu-Chen Hsu, Chia-Ming Hsieh, Sean-Lin Huang, Kuan-Hui Shih, and Ling-Ming Tseng. 2022. "Comparing Genetic Risk and Clinical Risk Classification in Luminal-like Breast Cancer Patients Using a 23-Gene Classifier" Cancers 14, no. 24: 6263. https://doi.org/10.3390/cancers14246263